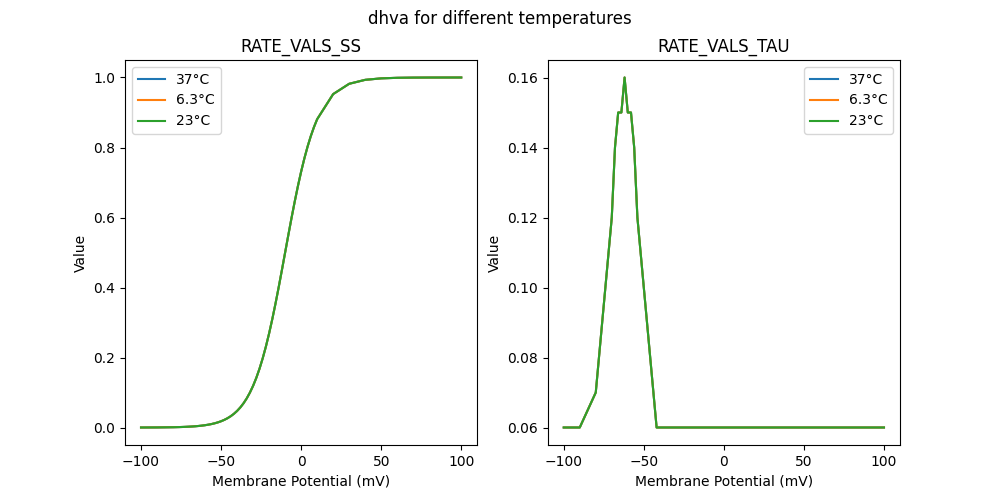
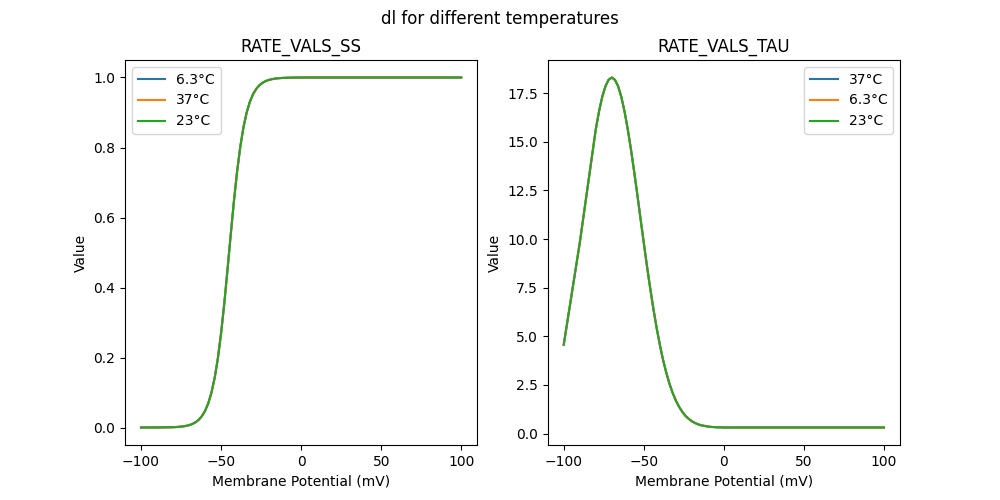
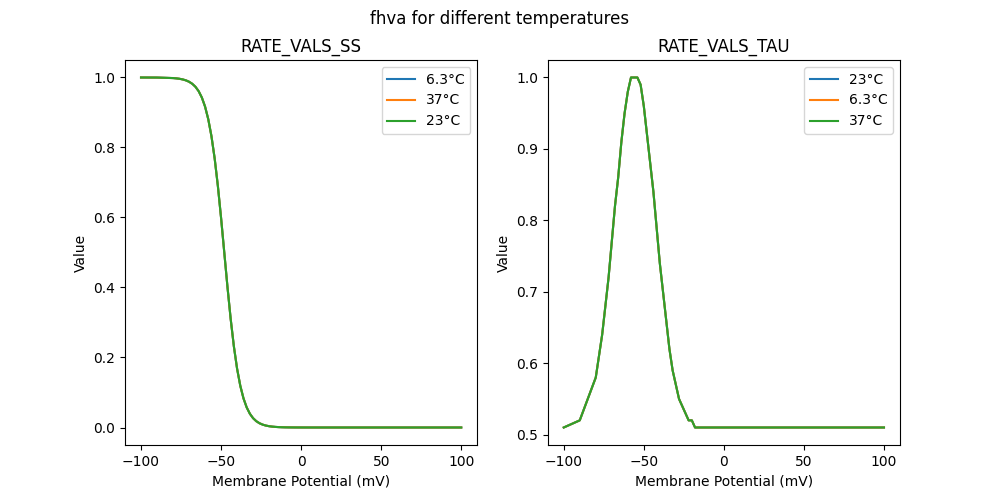
127507_newcachan.mod
Ion Channel Class: Ca
SUFFIX
cachan
GMAX NAME
gcalbar_cachan
G VALS
- gcahvabar_cachan: 0
- gcalbar_cachan: 0.000011196
I VALS
- 37: 0.0028331310910510205
- 6.3: 0.0028331310910510205
STATES
- dhva
- dl
- fhva
GATES
- dhva: 1
- dl: 1
- fhva: 1
ERROR FLAGS
- 8
RATES
true
Q10 TAU
1
Q10 G
1
RATE VALS TAU
{
"fhva_5e-05_23": [
0.51,
0.52,
0.58,
0.61,
0.64,
0.68,
0.72,
0.77,
0.82,
0.86,
0.91,
0.95,
0.98,
1,
1,
1,
0.99,
0.96,
0.92,
0.88,
0.84,
0.79,
0.74,
0.7,
0.66,
0.62,
0.59,
0.57,
0.55,
0.54,
0.53,
0.52,
0.52,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51
],
"fhva_5e-05_6.3": [
0.51,
0.52,
0.58,
0.61,
0.64,
0.68,
0.72,
0.77,
0.82,
0.86,
0.91,
0.95,
0.98,
1,
1,
1,
0.99,
0.96,
0.92,
0.88,
0.84,
0.79,
0.74,
0.7,
0.66,
0.62,
0.59,
0.57,
0.55,
0.54,
0.53,
0.52,
0.52,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51
],
"dhva_5e-05_37": [
0.06,
0.06,
0.07,
0.08,
0.09,
0.1,
0.11,
0.12,
0.14,
0.15,
0.15,
0.16,
0.15,
0.15,
0.14,
0.12,
0.11,
0.1,
0.09,
0.08,
0.07,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06
],
"dl_5e-05_37": [
4.57,
9.8,
15.64,
16.55,
17.3,
17.85,
18.19,
18.31,
18.19,
17.85,
17.3,
16.55,
15.64,
14.6,
13.46,
12.26,
11.02,
9.8,
8.6,
7.47,
6.41,
5.44,
4.57,
3.8,
3.14,
2.57,
2.09,
1.7,
1.38,
1.12,
0.91,
0.76,
0.63,
0.54,
0.47,
0.42,
0.39,
0.36,
0.34,
0.33,
0.32,
0.32,
0.31,
0.31,
0.31,
0.31,
0.31,
0.31,
0.31,
0.31,
0.31,
0.31,
0.31,
0.31,
0.31,
0.31
],
"dhva_5e-05_6.3": [
0.06,
0.06,
0.07,
0.08,
0.09,
0.1,
0.11,
0.12,
0.14,
0.15,
0.15,
0.16,
0.15,
0.15,
0.14,
0.12,
0.11,
0.1,
0.09,
0.08,
0.07,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06
],
"dl_5e-05_6.3": [
4.57,
9.8,
15.64,
16.55,
17.3,
17.85,
18.19,
18.31,
18.19,
17.85,
17.3,
16.55,
15.64,
14.6,
13.46,
12.26,
11.02,
9.8,
8.6,
7.47,
6.41,
5.44,
4.57,
3.8,
3.14,
2.57,
2.09,
1.7,
1.38,
1.12,
0.91,
0.76,
0.63,
0.54,
0.47,
0.42,
0.39,
0.36,
0.34,
0.33,
0.32,
0.32,
0.31,
0.31,
0.31,
0.31,
0.31,
0.31,
0.31,
0.31,
0.31,
0.31,
0.31,
0.31,
0.31,
0.31
],
"dhva_5e-05_23": [
0.06,
0.06,
0.07,
0.08,
0.09,
0.1,
0.11,
0.12,
0.14,
0.15,
0.15,
0.16,
0.15,
0.15,
0.14,
0.12,
0.11,
0.1,
0.09,
0.08,
0.07,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06
],
"fhva_5e-05_37": [
0.51,
0.52,
0.58,
0.61,
0.64,
0.68,
0.72,
0.77,
0.82,
0.86,
0.91,
0.95,
0.98,
1,
1,
1,
0.99,
0.96,
0.92,
0.88,
0.84,
0.79,
0.74,
0.7,
0.66,
0.62,
0.59,
0.57,
0.55,
0.54,
0.53,
0.52,
0.52,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51
],
"dl_5e-05_23": [
4.57,
9.8,
15.64,
16.55,
17.3,
17.85,
18.19,
18.31,
18.19,
17.85,
17.3,
16.55,
15.64,
14.6,
13.46,
12.26,
11.02,
9.8,
8.6,
7.47,
6.41,
5.44,
4.57,
3.8,
3.14,
2.57,
2.09,
1.7,
1.38,
1.12,
0.91,
0.76,
0.63,
0.54,
0.47,
0.42,
0.39,
0.36,
0.34,
0.33,
0.32,
0.32,
0.31,
0.31,
0.31,
0.31,
0.31,
0.31,
0.31,
0.31,
0.31,
0.31,
0.31,
0.31,
0.31,
0.31
]
}
RATE VALS SS
{
"dhva_5e-05_37": [
0.000123,
0.000335,
0.000911,
0.001113,
0.001359,
0.001659,
0.002025,
0.002473,
0.003018,
0.003684,
0.004496,
0.005486,
0.006693,
0.008163,
0.009952,
0.012128,
0.014774,
0.017986,
0.021881,
0.026597,
0.032295,
0.039166,
0.047426,
0.057324,
0.069138,
0.083172,
0.09975,
0.119202,
0.14185,
0.167981,
0.197815,
0.231474,
0.26894,
0.310024,
0.354342,
0.40131,
0.450164,
0.499998,
0.549832,
0.598685,
0.645654,
0.689972,
0.731057,
0.768523,
0.802182,
0.832017,
0.858148,
0.880796,
0.952574,
0.982014,
0.993307,
0.997527,
0.999089,
0.999665,
0.999877,
0.999983
],
"dl_5e-05_6.3": [
0.000017,
0.000123,
0.000911,
0.001359,
0.002025,
0.003018,
0.004496,
0.006693,
0.009952,
0.014774,
0.021881,
0.032296,
0.047426,
0.069139,
0.099751,
0.141851,
0.197816,
0.268942,
0.354343,
0.450165,
0.549833,
0.645656,
0.731058,
0.802183,
0.858148,
0.900249,
0.930861,
0.952574,
0.967704,
0.978118,
0.985226,
0.990048,
0.993307,
0.995504,
0.996982,
0.997975,
0.998641,
0.999089,
0.999389,
0.99959,
0.999725,
0.999816,
0.999877,
0.999917,
0.999945,
0.999963,
0.999975,
0.999983,
0.999998,
1,
1,
1,
1,
1,
1,
1
],
"dl_5e-05_37": [
0.000017,
0.000123,
0.000911,
0.001359,
0.002025,
0.003018,
0.004496,
0.006693,
0.009952,
0.014774,
0.021881,
0.032296,
0.047426,
0.069139,
0.099751,
0.141851,
0.197816,
0.268942,
0.354343,
0.450165,
0.549833,
0.645656,
0.731058,
0.802183,
0.858148,
0.900249,
0.930861,
0.952574,
0.967704,
0.978118,
0.985226,
0.990048,
0.993307,
0.995504,
0.996982,
0.997975,
0.998641,
0.999089,
0.999389,
0.99959,
0.999725,
0.999816,
0.999877,
0.999917,
0.999945,
0.999963,
0.999975,
0.999983,
0.999998,
1,
1,
1,
1,
1,
1,
1
],
"dhva_5e-05_6.3": [
0.000123,
0.000335,
0.000911,
0.001113,
0.001359,
0.001659,
0.002025,
0.002473,
0.003018,
0.003684,
0.004496,
0.005486,
0.006693,
0.008163,
0.009952,
0.012128,
0.014774,
0.017986,
0.021881,
0.026597,
0.032295,
0.039166,
0.047426,
0.057324,
0.069138,
0.083172,
0.09975,
0.119202,
0.14185,
0.167981,
0.197815,
0.231474,
0.26894,
0.310024,
0.354342,
0.40131,
0.450164,
0.499998,
0.549832,
0.598685,
0.645654,
0.689972,
0.731057,
0.768523,
0.802182,
0.832017,
0.858148,
0.880796,
0.952574,
0.982014,
0.993307,
0.997527,
0.999089,
0.999665,
0.999877,
0.999983
],
"dhva_5e-05_23": [
0.000123,
0.000335,
0.000911,
0.001113,
0.001359,
0.001659,
0.002025,
0.002473,
0.003018,
0.003684,
0.004496,
0.005486,
0.006693,
0.008163,
0.009952,
0.012128,
0.014774,
0.017986,
0.021881,
0.026597,
0.032295,
0.039166,
0.047426,
0.057324,
0.069138,
0.083172,
0.09975,
0.119202,
0.14185,
0.167981,
0.197815,
0.231474,
0.26894,
0.310024,
0.354342,
0.40131,
0.450164,
0.499998,
0.549832,
0.598685,
0.645654,
0.689972,
0.731057,
0.768523,
0.802182,
0.832017,
0.858148,
0.880796,
0.952574,
0.982014,
0.993307,
0.997527,
0.999089,
0.999665,
0.999877,
0.999983
],
"dl_5e-05_23": [
0.000017,
0.000123,
0.000911,
0.001359,
0.002025,
0.003018,
0.004496,
0.006693,
0.009952,
0.014774,
0.021881,
0.032296,
0.047426,
0.069139,
0.099751,
0.141851,
0.197816,
0.268942,
0.354343,
0.450165,
0.549833,
0.645656,
0.731058,
0.802183,
0.858148,
0.900249,
0.930861,
0.952574,
0.967704,
0.978118,
0.985226,
0.990048,
0.993307,
0.995504,
0.996982,
0.997975,
0.998641,
0.999089,
0.999389,
0.99959,
0.999725,
0.999816,
0.999877,
0.999917,
0.999945,
0.999963,
0.999975,
0.999983,
0.999998,
1,
1,
1,
1,
1,
1,
1
],
"fhva_5e-05_6.3": [
0.99997,
0.999775,
0.998341,
0.997527,
0.996316,
0.994514,
0.991837,
0.987872,
0.982014,
0.973403,
0.960834,
0.942676,
0.916827,
0.880797,
0.832019,
0.768525,
0.689975,
0.598689,
0.500001,
0.401314,
0.310027,
0.231476,
0.167983,
0.119204,
0.083173,
0.057325,
0.039166,
0.026597,
0.017986,
0.012129,
0.008163,
0.005486,
0.003684,
0.002473,
0.001659,
0.001113,
0.000746,
0.0005,
0.000335,
0.000225,
0.000151,
0.000101,
0.000068,
0.000045,
0.00003,
0.00002,
0.000014,
0.000009,
0.000001,
0,
0,
0,
0,
0,
0,
0
],
"fhva_5e-05_37": [
0.99997,
0.999775,
0.998341,
0.997527,
0.996316,
0.994514,
0.991837,
0.987872,
0.982014,
0.973403,
0.960834,
0.942676,
0.916827,
0.880797,
0.832019,
0.768525,
0.689975,
0.598689,
0.500001,
0.401314,
0.310027,
0.231476,
0.167983,
0.119204,
0.083173,
0.057325,
0.039166,
0.026597,
0.017986,
0.012129,
0.008163,
0.005486,
0.003684,
0.002473,
0.001659,
0.001113,
0.000746,
0.0005,
0.000335,
0.000225,
0.000151,
0.000101,
0.000068,
0.000045,
0.00003,
0.00002,
0.000014,
0.000009,
0.000001,
0,
0,
0,
0,
0,
0,
0
],
"fhva_5e-05_23": [
0.99997,
0.999775,
0.998341,
0.997527,
0.996316,
0.994514,
0.991837,
0.987872,
0.982014,
0.973403,
0.960834,
0.942676,
0.916827,
0.880797,
0.832019,
0.768525,
0.689975,
0.598689,
0.500001,
0.401314,
0.310027,
0.231476,
0.167983,
0.119204,
0.083173,
0.057325,
0.039166,
0.026597,
0.017986,
0.012129,
0.008163,
0.005486,
0.003684,
0.002473,
0.001659,
0.001113,
0.000746,
0.0005,
0.000335,
0.000225,
0.000151,
0.000101,
0.000068,
0.000045,
0.00003,
0.00002,
0.000014,
0.000009,
0.000001,
0,
0,
0,
0,
0,
0,
0
]
}
SM1 FIT
false
SM2 FIT
false
SM3 FIT
false
SM4 FIT
false
SM5 FIT
false
Figures