20212_My_cal.mod
Ion Channel Class: Ca
SUFFIX
mycal
GMAX NAME
gcalbar_mycal
G VALS
- gcalbar_mycal: 1
I VALS
{}
STATES
- m
- h
GATES
- m: 2
- h: 0
ERROR FLAGS
- 4
RATES
true
RATE VALS TAU
{
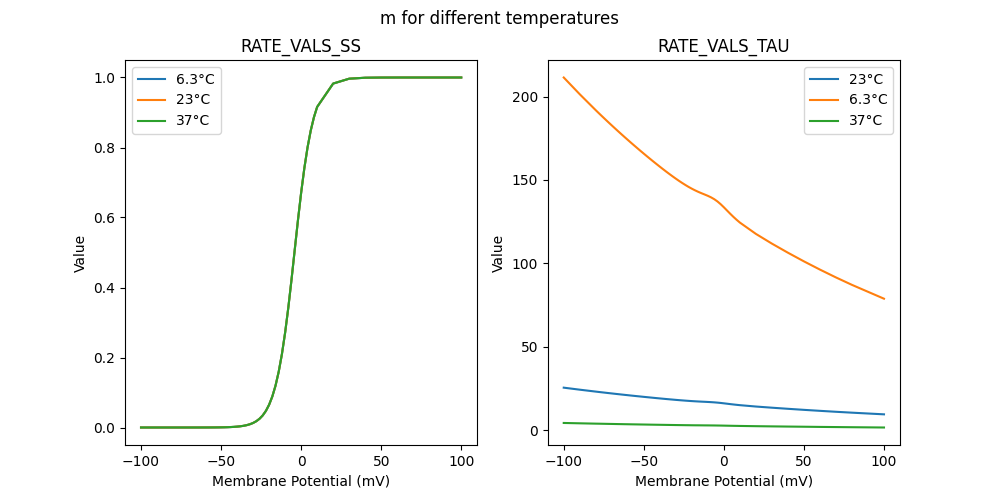
"m_5e-05_23": [
25.5,
24.29,
23.14,
22.91,
22.69,
22.47,
22.25,
22.04,
21.82,
21.61,
21.4,
21.19,
20.99,
20.78,
20.58,
20.38,
20.19,
19.99,
19.8,
19.61,
19.42,
19.23,
19.05,
18.87,
18.69,
18.52,
18.35,
18.18,
18.02,
17.87,
17.72,
17.58,
17.45,
17.33,
17.22,
17.12,
17.03,
16.93,
16.82,
16.69,
16.53,
16.34,
16.11,
15.88,
15.64,
15.42,
15.21,
15.01,
14.19,
13.49,
12.83,
12.2,
11.61,
11.04,
10.51,
9.51
],
"m_5e-05_6.3": [
211.43,
201.39,
191.82,
189.96,
188.12,
186.3,
184.49,
182.71,
180.94,
179.18,
177.45,
175.73,
174.03,
172.34,
170.68,
169.03,
167.39,
165.78,
164.18,
162.6,
161.04,
159.5,
157.98,
156.48,
155.01,
153.57,
152.16,
150.79,
149.46,
148.19,
146.98,
145.83,
144.77,
143.79,
142.89,
142.07,
141.29,
140.49,
139.58,
138.48,
137.12,
135.48,
133.63,
131.67,
129.72,
127.85,
126.1,
124.47,
117.68,
111.85,
106.4,
101.21,
96.28,
91.58,
87.12,
78.83
],
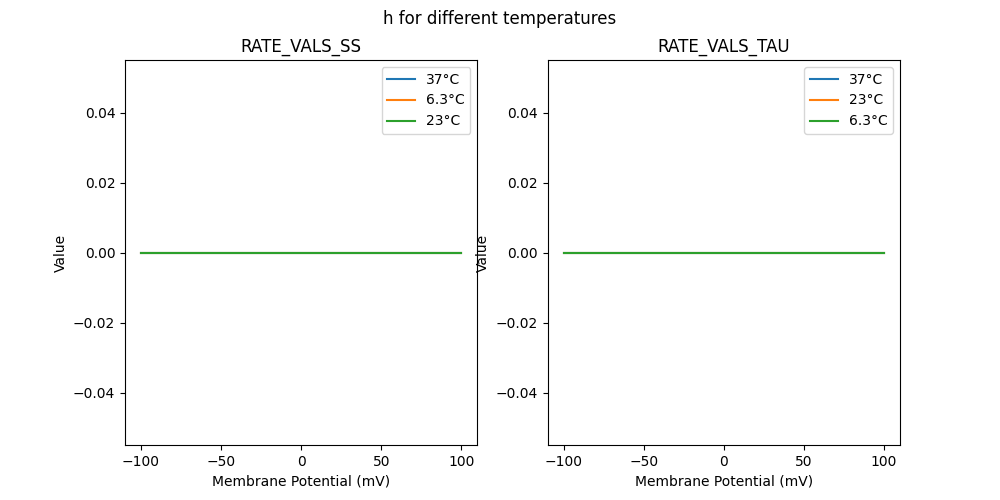
"h_5e-05_37": [
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0
],
"h_5e-05_23": [
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0
],
"m_5e-05_37": [
4.34,
4.13,
3.93,
3.9,
3.86,
3.82,
3.78,
3.75,
3.71,
3.67,
3.64,
3.6,
3.57,
3.53,
3.5,
3.47,
3.43,
3.4,
3.37,
3.33,
3.3,
3.27,
3.24,
3.21,
3.18,
3.15,
3.12,
3.09,
3.06,
3.04,
3.01,
2.99,
2.96,
2.94,
2.93,
2.91,
2.89,
2.88,
2.86,
2.84,
2.81,
2.78,
2.74,
2.7,
2.66,
2.62,
2.59,
2.55,
2.41,
2.29,
2.18,
2.08,
1.97,
1.88,
1.79,
1.62
],
"h_5e-05_6.3": [
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0
]
}
RATE VALS SS
{
"h_5e-05_37": [
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0
],
"m_5e-05_6.3": [
0,
0.000001,
0.000003,
0.000004,
0.000006,
0.000009,
0.000012,
0.000017,
0.000023,
0.000033,
0.000045,
0.000063,
0.000088,
0.000123,
0.000172,
0.00024,
0.000335,
0.000468,
0.000653,
0.000911,
0.001271,
0.001773,
0.002473,
0.003448,
0.004805,
0.006694,
0.009317,
0.012956,
0.01799,
0.024931,
0.034459,
0.047454,
0.06503,
0.088534,
0.119492,
0.15947,
0.209794,
0.271124,
0.342927,
0.423047,
0.507655,
0.591867,
0.670677,
0.740747,
0.800033,
0.848274,
0.886356,
0.915718,
0.982561,
0.996527,
0.999257,
0.99979,
0.99989,
0.999917,
0.999918,
0.999928
],
"h_5e-05_6.3": [
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0
],
"m_5e-05_23": [
0,
0.000001,
0.000003,
0.000004,
0.000006,
0.000009,
0.000012,
0.000017,
0.000023,
0.000033,
0.000045,
0.000063,
0.000088,
0.000123,
0.000172,
0.00024,
0.000335,
0.000468,
0.000653,
0.000911,
0.001271,
0.001773,
0.002473,
0.003447,
0.004805,
0.006693,
0.009316,
0.012954,
0.017988,
0.024928,
0.034454,
0.047447,
0.06502,
0.088519,
0.119469,
0.159434,
0.209741,
0.271048,
0.342825,
0.422925,
0.507532,
0.591761,
0.670759,
0.740884,
0.800216,
0.848487,
0.886573,
0.915929,
0.982695,
0.996631,
0.99935,
0.999874,
0.999972,
0.999989,
0.999995,
0.999997
],
"h_5e-05_23": [
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0
],
"m_5e-05_37": [
0,
0.000001,
0.000003,
0.000004,
0.000006,
0.000009,
0.000012,
0.000017,
0.000023,
0.000033,
0.000045,
0.000063,
0.000088,
0.000123,
0.000172,
0.00024,
0.000335,
0.000468,
0.000653,
0.000911,
0.001271,
0.001773,
0.002473,
0.003447,
0.004805,
0.006693,
0.009316,
0.012954,
0.017987,
0.024928,
0.034453,
0.047446,
0.065019,
0.088515,
0.119462,
0.159422,
0.209719,
0.271013,
0.342776,
0.422867,
0.507478,
0.591727,
0.670769,
0.740932,
0.800293,
0.848581,
0.886672,
0.916023,
0.982734,
0.996642,
0.999357,
0.999878,
0.999977,
0.999996,
0.999999,
1
]
}
SM1 FIT
false
SM2 FIT
false
SM3 FIT
false
SM4 FIT
false
SM5 FIT
false
Figures