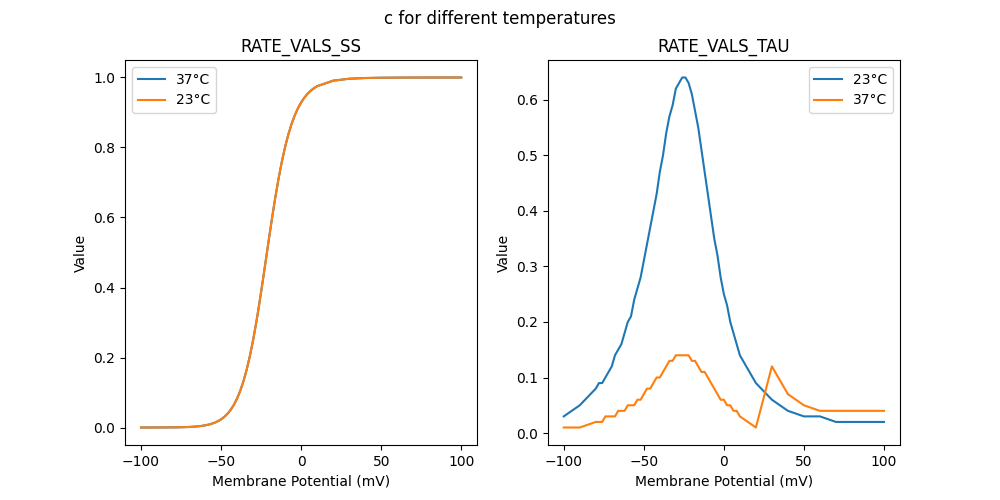
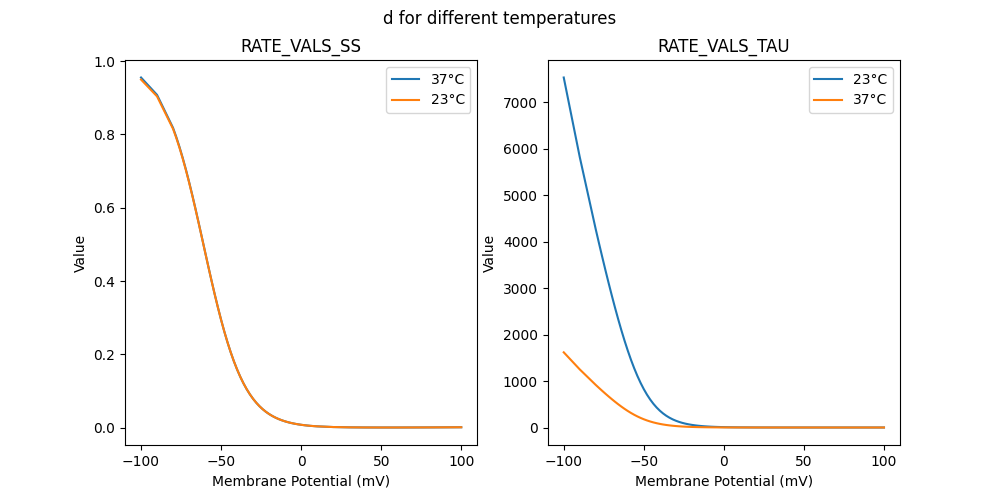
51781_nca.mod
Ion Channel Class: Ca
SUFFIX
nca
GMAX NAME
gncabar_nca
G VALS
- gncabar_nca: 1
I VALS
- 37: 0.9999568690252919
- 6.3: 0.9999794047670708
STATES
- c
- d
GATES
- c: 2
- d: 1
ERROR FLAGS
- 19
RATES
true
RATE VALS TAU
{
"c_5e-05_23": [
0.03,
0.05,
0.08,
0.09,
0.09,
0.1,
0.11,
0.12,
0.14,
0.15,
0.16,
0.18,
0.2,
0.21,
0.24,
0.26,
0.28,
0.31,
0.34,
0.37,
0.4,
0.43,
0.47,
0.5,
0.54,
0.57,
0.59,
0.62,
0.63,
0.64,
0.64,
0.63,
0.61,
0.58,
0.55,
0.51,
0.47,
0.43,
0.39,
0.35,
0.32,
0.28,
0.25,
0.23,
0.2,
0.18,
0.16,
0.14,
0.09,
0.06,
0.04,
0.03,
0.03,
0.02,
0.02,
0.02
],
"c_5e-05_37": [
0.01,
0.01,
0.02,
0.02,
0.02,
0.03,
0.03,
0.03,
0.03,
0.04,
0.04,
0.04,
0.05,
0.05,
0.05,
0.06,
0.06,
0.07,
0.08,
0.08,
0.09,
0.1,
0.1,
0.11,
0.12,
0.13,
0.13,
0.14,
0.14,
0.14,
0.14,
0.14,
0.13,
0.13,
0.12,
0.11,
0.11,
0.1,
0.09,
0.08,
0.07,
0.06,
0.06,
0.05,
0.05,
0.04,
0.04,
0.03,
0.01,
0.12,
0.07,
0.05,
0.04,
0.04,
0.04,
0.04
],
"d_5e-05_23": [
7535.84,
5823.32,
4265.7,
3969.2,
3678.12,
3393.06,
3114.87,
2844.57,
2583.34,
2332.46,
2093.25,
1866.96,
1654.71,
1457.42,
1275.72,
1109.95,
960.11,
825.9,
706.75,
601.86,
510.23,
430.78,
362.35,
303.77,
253.9,
211.65,
176.02,
146.09,
121.03,
100.11,
82.7,
68.24,
56.26,
46.34,
38.15,
31.39,
25.81,
21.23,
17.45,
14.35,
11.8,
9.71,
7.99,
6.58,
5.42,
4.47,
3.69,
3.05,
1.23,
0.56,
0.31,
0.22,
0.18,
0.17,
0.17,
0.16
],
"d_5e-05_37": [
1618.69,
1250.84,
916.27,
852.58,
790.06,
728.83,
669.08,
611.02,
554.91,
501.02,
449.64,
401.03,
355.44,
313.06,
274.03,
238.42,
206.24,
177.41,
151.82,
129.29,
109.6,
92.54,
77.84,
65.26,
54.54,
45.47,
37.81,
31.38,
26,
21.51,
17.77,
14.66,
12.09,
9.96,
8.2,
6.75,
5.55,
4.56,
3.75,
3.09,
2.54,
2.09,
1.72,
1.42,
1.17,
0.96,
0.8,
0.66,
0.27,
0.12,
0.07,
0.05,
0.04,
0.04,
0.04,
0.04
]
}
RATE VALS SS
{
"d_5e-05_37": [
0.955092,
0.907764,
0.817842,
0.793153,
0.766037,
0.736534,
0.704711,
0.670746,
0.634884,
0.597446,
0.558824,
0.519465,
0.479858,
0.440493,
0.401858,
0.364401,
0.328516,
0.294528,
0.262677,
0.233132,
0.205982,
0.181247,
0.158889,
0.138822,
0.120927,
0.105059,
0.091059,
0.078762,
0.068005,
0.058625,
0.050472,
0.043404,
0.03729,
0.032012,
0.027464,
0.02355,
0.020186,
0.017298,
0.014822,
0.0127,
0.010883,
0.009328,
0.007999,
0.006863,
0.005893,
0.005066,
0.004359,
0.003758,
0.001856,
0.001028,
0.000696,
0.000599,
0.00062,
0.00071,
0.000849,
0.001264
],
"c_5e-05_37": [
0.000025,
0.0001,
0.0004,
0.000527,
0.000694,
0.000914,
0.001202,
0.001582,
0.002079,
0.002731,
0.003585,
0.004703,
0.006162,
0.008067,
0.010547,
0.013769,
0.017946,
0.023342,
0.030289,
0.039188,
0.050525,
0.064865,
0.082846,
0.105155,
0.13248,
0.165437,
0.204465,
0.249707,
0.300891,
0.357243,
0.417479,
0.47989,
0.542522,
0.603418,
0.660855,
0.713519,
0.760591,
0.801733,
0.83701,
0.86678,
0.891579,
0.912023,
0.928743,
0.942337,
0.95334,
0.96222,
0.969375,
0.975134,
0.990913,
0.996431,
0.998489,
0.999315,
0.999672,
0.999836,
0.999915,
0.999976
],
"d_5e-05_23": [
0.949486,
0.90364,
0.815111,
0.790687,
0.763835,
0.734578,
0.702993,
0.669255,
0.633602,
0.596353,
0.557906,
0.518705,
0.479235,
0.439989,
0.401458,
0.364086,
0.328271,
0.294336,
0.26253,
0.233022,
0.205901,
0.181187,
0.158844,
0.13879,
0.120903,
0.105041,
0.091046,
0.078753,
0.067998,
0.058621,
0.050469,
0.043402,
0.037289,
0.032011,
0.027463,
0.023549,
0.020185,
0.017298,
0.014821,
0.012699,
0.010883,
0.009328,
0.007999,
0.006863,
0.005893,
0.005066,
0.004359,
0.003758,
0.001856,
0.001028,
0.000696,
0.000599,
0.00062,
0.00071,
0.000849,
0.001264
],
"c_5e-05_23": [
0.000025,
0.0001,
0.0004,
0.000527,
0.000694,
0.000914,
0.001202,
0.001582,
0.002079,
0.002731,
0.003585,
0.004703,
0.006162,
0.008067,
0.010547,
0.013769,
0.017946,
0.023342,
0.030289,
0.039188,
0.050525,
0.064865,
0.082846,
0.105155,
0.13248,
0.165437,
0.204465,
0.249707,
0.300891,
0.357243,
0.417479,
0.47989,
0.542522,
0.603418,
0.660855,
0.713519,
0.760591,
0.801733,
0.83701,
0.86678,
0.891579,
0.912023,
0.928743,
0.942337,
0.95334,
0.96222,
0.969375,
0.975134,
0.990913,
0.996431,
0.998489,
0.999315,
0.999672,
0.999836,
0.999915,
0.999976
]
}
SM1 FIT
false
SM2 FIT
false
SM3 FIT
false
SM4 FIT
false
SM5 FIT
false
Figures