150621_KAsm.mod
Ion Channel Class: K
SUFFIX
KAsm
GMAX NAME
gkasmbar_KAsm
G VALS
- gkasmbar_KAsm: 0.00032
I VALS
- 37: 0.0003200003634935401
- 6.3: 0.0003200002395565714
STATES
- m
- h
GATES
- m: 1
- h: 1
ERROR FLAGS
- 19
RATES
true
RATE VALS TAU
{
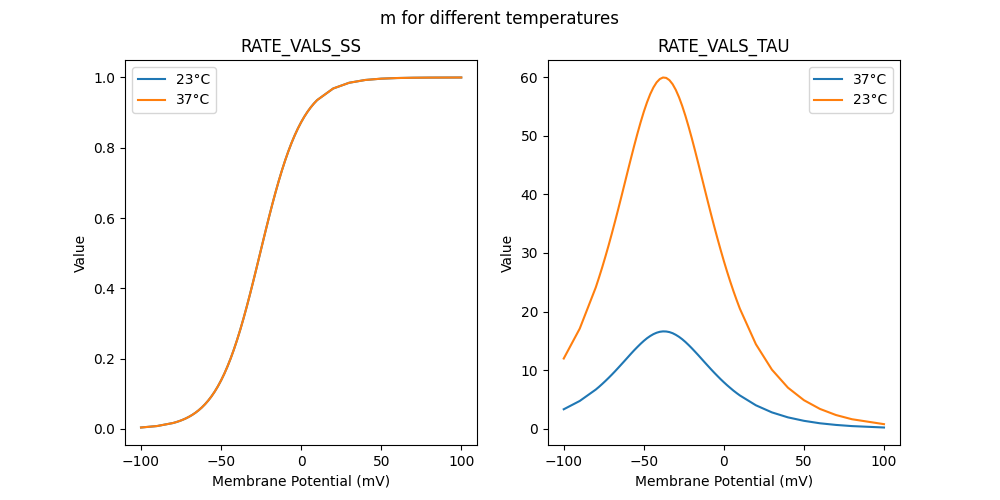
"m_37": [
3.33,
4.75,
6.69,
7.15,
7.64,
8.15,
8.68,
9.23,
9.8,
10.39,
10.99,
11.59,
12.2,
12.81,
13.4,
13.96,
14.5,
15,
15.45,
15.83,
16.15,
16.39,
16.55,
16.62,
16.6,
16.5,
16.31,
16.03,
15.69,
15.28,
14.81,
14.29,
13.74,
13.16,
12.57,
11.96,
11.35,
10.75,
10.15,
9.57,
9.01,
8.46,
7.94,
7.44,
6.97,
6.51,
6.09,
5.69,
4.01,
2.8,
1.95,
1.36,
0.94,
0.66,
0.46,
0.22
],
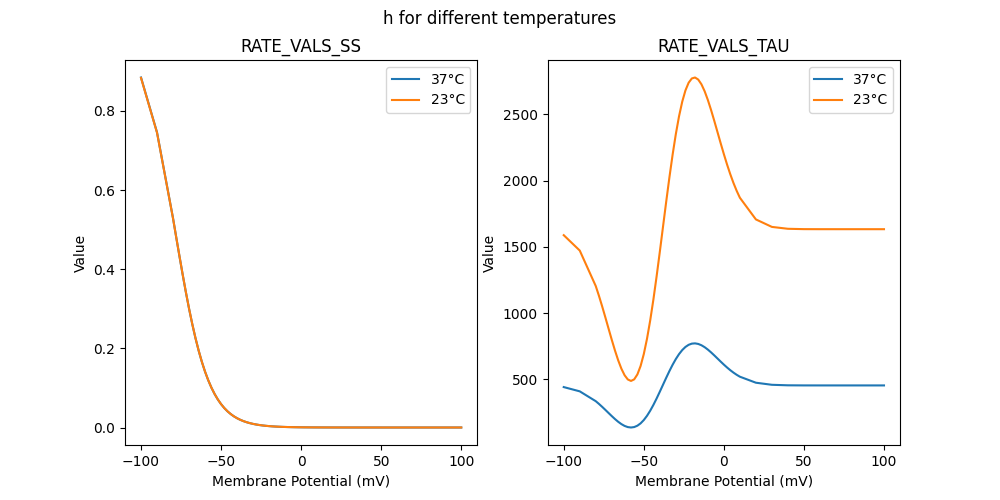
"h_37": [
440.31,
408.1,
333.69,
313.14,
291.11,
268.03,
244.46,
221.08,
198.69,
178.22,
160.64,
146.93,
138.07,
134.94,
138.28,
148.64,
166.3,
191.3,
223.32,
261.77,
305.75,
354.08,
405.39,
458.14,
510.72,
561.55,
609.12,
652.08,
689.31,
719.96,
743.51,
759.7,
768.62,
770.62,
766.28,
756.37,
741.81,
723.57,
702.64,
679.99,
656.51,
632.98,
610.07,
588.31,
568.09,
549.67,
533.21,
518.75,
473.33,
457.63,
453.69,
452.96,
452.85,
452.84,
452.84,
452.84
],
"h_23": [
1588.07,
1471.89,
1203.53,
1129.4,
1049.96,
966.72,
881.69,
797.35,
716.63,
642.79,
579.36,
529.93,
497.97,
486.68,
498.73,
536.08,
599.8,
689.94,
805.45,
944.14,
1102.75,
1277.07,
1462.12,
1652.37,
1842.04,
2025.37,
2196.93,
2351.87,
2486.15,
2596.72,
2681.63,
2740.04,
2772.21,
2779.41,
2763.76,
2728.04,
2675.52,
2609.72,
2534.23,
2452.53,
2367.84,
2282.98,
2200.35,
2121.87,
2048.94,
1982.52,
1923.14,
1870.97,
1707.15,
1650.54,
1636.34,
1633.69,
1633.32,
1633.28,
1633.28,
1633.28
],
"m_23": [
11.99,
17.1,
24.12,
25.79,
27.53,
29.37,
31.28,
33.28,
35.34,
37.45,
39.61,
41.8,
44,
46.17,
48.3,
50.35,
52.29,
54.09,
55.7,
57.1,
58.24,
59.11,
59.68,
59.94,
59.87,
59.49,
58.8,
57.82,
56.57,
55.08,
53.39,
51.53,
49.55,
47.46,
45.31,
43.12,
40.93,
38.75,
36.6,
34.5,
32.47,
30.51,
28.62,
26.82,
25.11,
23.48,
21.95,
20.49,
14.43,
10.09,
7.02,
4.88,
3.39,
2.35,
1.63,
0.79
]
}
RATE VALS SS
{
"m_23": [
0.003706,
0.007829,
0.01646,
0.01908,
0.022108,
0.025603,
0.029635,
0.034279,
0.039621,
0.045756,
0.052789,
0.060834,
0.070014,
0.080462,
0.092313,
0.105709,
0.120791,
0.137693,
0.156538,
0.177433,
0.200454,
0.225642,
0.252994,
0.282451,
0.313898,
0.347152,
0.381967,
0.418038,
0.455008,
0.492481,
0.530038,
0.567258,
0.603734,
0.639091,
0.673003,
0.705198,
0.735469,
0.763673,
0.78973,
0.813616,
0.835353,
0.855008,
0.872673,
0.888467,
0.902521,
0.914973,
0.925966,
0.935637,
0.968585,
0.984939,
0.992842,
0.996612,
0.9984,
0.999245,
0.999644,
0.999921
],
"h_37": [
0.884391,
0.745613,
0.528643,
0.480634,
0.432974,
0.38672,
0.342204,
0.300305,
0.261496,
0.226086,
0.194209,
0.165865,
0.140936,
0.119218,
0.100456,
0.084365,
0.07065,
0.05902,
0.049205,
0.040951,
0.034031,
0.028247,
0.023422,
0.019405,
0.016066,
0.013293,
0.010994,
0.009088,
0.00751,
0.006205,
0.005125,
0.004232,
0.003494,
0.002885,
0.002381,
0.001966,
0.001622,
0.001339,
0.001105,
0.000912,
0.000752,
0.000621,
0.000512,
0.000423,
0.000349,
0.000288,
0.000237,
0.000196,
0.000075,
0.000029,
0.000011,
0.000004,
0.000002,
0.000001,
0,
0
],
"m_37": [
0.003706,
0.007829,
0.01646,
0.01908,
0.022108,
0.025603,
0.029635,
0.034279,
0.039621,
0.045756,
0.052789,
0.060834,
0.070014,
0.080462,
0.092313,
0.105709,
0.120791,
0.137693,
0.156538,
0.177433,
0.200454,
0.225642,
0.252994,
0.282451,
0.313898,
0.347152,
0.381967,
0.418038,
0.455008,
0.492481,
0.530038,
0.567258,
0.603734,
0.639091,
0.673003,
0.705198,
0.735469,
0.763673,
0.78973,
0.813616,
0.835353,
0.855008,
0.872673,
0.888467,
0.902521,
0.914973,
0.925966,
0.935637,
0.968585,
0.984939,
0.992842,
0.996612,
0.9984,
0.999245,
0.999644,
0.999921
],
"h_23": [
0.883383,
0.744822,
0.528181,
0.480241,
0.433546,
0.38699,
0.342422,
0.300477,
0.261632,
0.226189,
0.19429,
0.165929,
0.140986,
0.11926,
0.100493,
0.084398,
0.07068,
0.05905,
0.049233,
0.040978,
0.034059,
0.028273,
0.023447,
0.019428,
0.016087,
0.013313,
0.011011,
0.009104,
0.007524,
0.006217,
0.005135,
0.004241,
0.003501,
0.002891,
0.002386,
0.001969,
0.001625,
0.001341,
0.001107,
0.000913,
0.000754,
0.000622,
0.000513,
0.000423,
0.000349,
0.000288,
0.000238,
0.000196,
0.000075,
0.000029,
0.000011,
0.000004,
0.000002,
0.000001,
0,
0
]
}
SM1 FIT
false
SM2 FIT
false
SM3 FIT
false
SM4 FIT
false
SM5 FIT
false
Figures