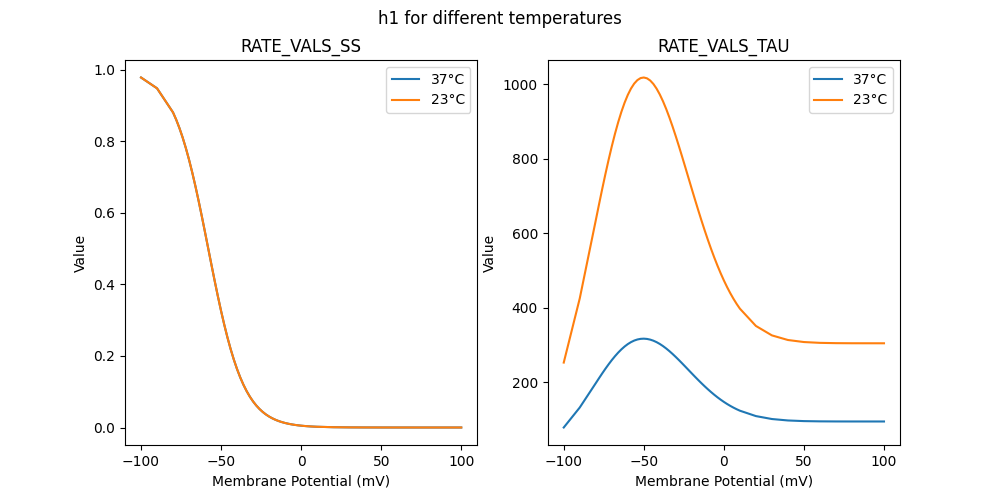
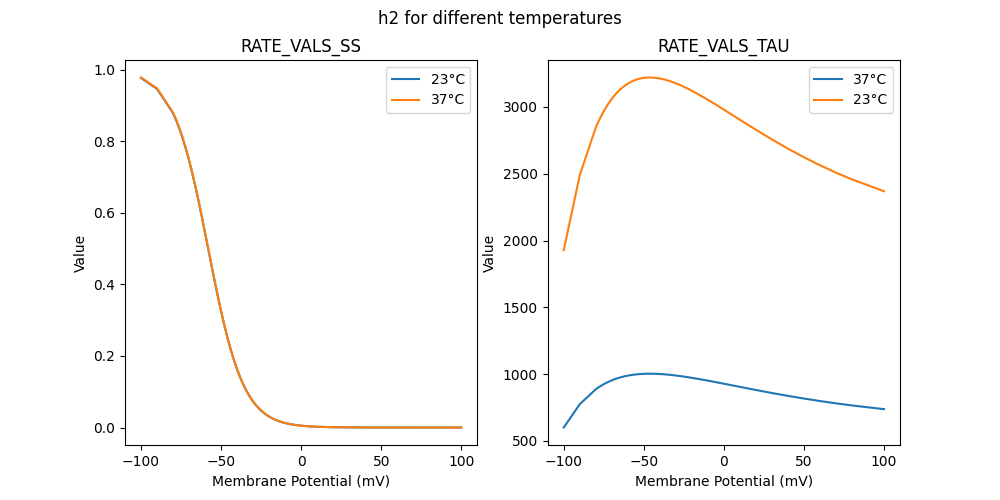
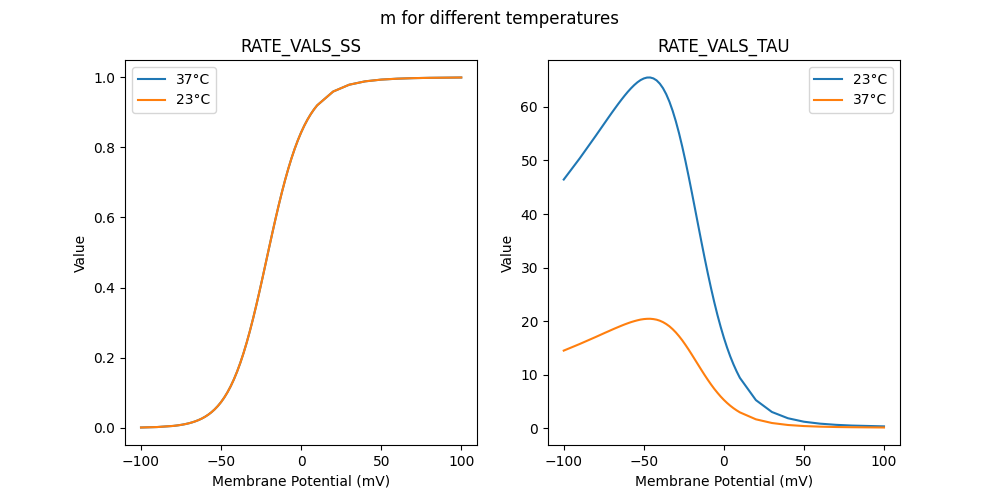
184180_Kv2like.mod
Ion Channel Class: K
SUFFIX
Kv2like
GMAX NAME
gbar_Kv2like
G VALS
- gbar_Kv2like: 0.00001
I VALS
- 37: 0.000015292133387919645
- 6.3: 0.000015292133416711217
STATES
- m
- h1
- h2
GATES
- m: 2
- h1: 1
- h2: 1
ERROR FLAGS
- 19
RATES
true
RATE VALS TAU
{
"h1_37": [
78.84,
132.48,
197.72,
210.89,
223.79,
236.29,
248.25,
259.56,
270.09,
279.75,
288.43,
296.06,
302.57,
307.91,
312.05,
314.97,
316.66,
317.15,
316.45,
314.62,
311.71,
307.79,
302.94,
297.24,
290.79,
283.68,
276.01,
267.89,
259.41,
250.67,
241.77,
232.79,
223.82,
214.93,
206.2,
197.68,
189.44,
181.5,
173.91,
166.7,
159.9,
153.5,
147.54,
141.99,
136.87,
132.16,
127.85,
123.93,
109.47,
101.58,
97.71,
96,
95.32,
95.08,
95,
94.97
],
"h2_37": [
600.83,
775.29,
886.87,
903.33,
918.14,
931.4,
943.23,
953.71,
962.94,
971.02,
978.01,
984,
989.06,
993.25,
996.63,
999.28,
1001.23,
1002.55,
1003.28,
1003.46,
1003.15,
1002.37,
1001.17,
999.58,
997.64,
995.37,
992.8,
989.96,
986.88,
983.57,
980.07,
976.38,
972.53,
968.54,
964.42,
960.19,
955.86,
951.44,
946.96,
942.41,
937.81,
933.17,
928.5,
923.8,
919.09,
914.37,
909.65,
904.94,
881.6,
859.05,
837.63,
817.54,
798.86,
781.64,
765.84,
738.29
],
"h1_23": [
253,
425.15,
634.54,
676.81,
718.21,
758.32,
796.72,
833.01,
866.82,
897.8,
925.67,
950.16,
971.05,
988.2,
1001.48,
1010.84,
1016.27,
1017.83,
1015.6,
1009.73,
1000.39,
987.81,
972.24,
953.95,
933.24,
910.42,
885.82,
859.75,
832.53,
804.49,
775.91,
747.1,
718.31,
689.79,
661.77,
634.43,
607.96,
582.48,
558.14,
535,
513.16,
492.64,
473.48,
455.69,
439.25,
424.14,
410.31,
397.73,
351.32,
325.98,
313.58,
308.1,
305.92,
305.13,
304.87,
304.77
],
"m_23": [
46.44,
50.41,
54.62,
55.48,
56.34,
57.2,
58.06,
58.91,
59.74,
60.56,
61.35,
62.11,
62.83,
63.49,
64.09,
64.61,
65.03,
65.33,
65.5,
65.51,
65.34,
64.97,
64.36,
63.51,
62.39,
60.99,
59.3,
57.33,
55.1,
52.61,
49.91,
47.03,
44.03,
40.95,
37.85,
34.77,
31.76,
28.87,
26.12,
23.53,
21.12,
18.91,
16.88,
15.04,
13.38,
11.9,
10.57,
9.38,
5.22,
3,
1.83,
1.18,
0.82,
0.6,
0.46,
0.31
],
"m_37": [
14.47,
15.71,
17.02,
17.29,
17.56,
17.83,
18.09,
18.36,
18.62,
18.87,
19.12,
19.36,
19.58,
19.79,
19.97,
20.13,
20.27,
20.36,
20.41,
20.42,
20.36,
20.25,
20.06,
19.79,
19.44,
19.01,
18.48,
17.87,
17.17,
16.4,
15.55,
14.66,
13.72,
12.76,
11.8,
10.84,
9.9,
9,
8.14,
7.33,
6.59,
5.89,
5.26,
4.69,
4.17,
3.71,
3.3,
2.93,
1.63,
0.94,
0.57,
0.37,
0.26,
0.19,
0.15,
0.1
],
"h2_23": [
1928.28,
2488.16,
2846.27,
2899.1,
2946.62,
2989.19,
3027.14,
3060.79,
3090.43,
3116.34,
3138.78,
3158.01,
3174.23,
3187.68,
3198.55,
3207.03,
3213.3,
3217.53,
3219.87,
3220.46,
3219.45,
3216.96,
3213.11,
3208.01,
3201.77,
3194.48,
3186.24,
3177.14,
3167.24,
3156.63,
3145.38,
3133.55,
3121.2,
3108.39,
3095.17,
3081.58,
3067.69,
3053.52,
3039.12,
3024.52,
3009.76,
2994.87,
2979.87,
2964.81,
2949.69,
2934.54,
2919.4,
2904.26,
2829.38,
2757.01,
2688.25,
2623.76,
2563.83,
2508.55,
2457.85,
2369.43
]
}
RATE VALS SS
{
"h2_23": [
0.976629,
0.945955,
0.87831,
0.857868,
0.834588,
0.808292,
0.778861,
0.746278,
0.710626,
0.672118,
0.631108,
0.588072,
0.54361,
0.498416,
0.453226,
0.408775,
0.365749,
0.324751,
0.286268,
0.250643,
0.218092,
0.188698,
0.162441,
0.139208,
0.118825,
0.101073,
0.085716,
0.072503,
0.06119,
0.051544,
0.043348,
0.036405,
0.030539,
0.025592,
0.02143,
0.017931,
0.014995,
0.012534,
0.010472,
0.008747,
0.007303,
0.006097,
0.005119,
0.004271,
0.003564,
0.002973,
0.00248,
0.002068,
0.000834,
0.000336,
0.000136,
0.000055,
0.000022,
0.000009,
0.000004,
0.000001
],
"m_37": [
0.000851,
0.002133,
0.005304,
0.006357,
0.007615,
0.009117,
0.010909,
0.013044,
0.015586,
0.018609,
0.022196,
0.026449,
0.031478,
0.037414,
0.044401,
0.052601,
0.062193,
0.073369,
0.086332,
0.101291,
0.118455,
0.138019,
0.160154,
0.184993,
0.212609,
0.243005,
0.276092,
0.311685,
0.349489,
0.389109,
0.430058,
0.471785,
0.513695,
0.555191,
0.595705,
0.634724,
0.67182,
0.706662,
0.739018,
0.768757,
0.795835,
0.820286,
0.842202,
0.86172,
0.879006,
0.894243,
0.907621,
0.919327,
0.958731,
0.978157,
0.987801,
0.992741,
0.995392,
0.996894,
0.997796,
0.998751
],
"h2_37": [
0.977924,
0.94757,
0.880028,
0.859583,
0.836276,
0.809947,
0.780477,
0.74785,
0.712135,
0.67356,
0.632469,
0.589344,
0.544795,
0.49951,
0.454224,
0.409675,
0.366557,
0.325472,
0.286902,
0.251199,
0.218574,
0.189115,
0.162799,
0.139515,
0.119086,
0.101296,
0.085904,
0.072661,
0.061323,
0.051656,
0.043442,
0.036484,
0.030604,
0.025647,
0.021475,
0.017969,
0.015027,
0.01256,
0.010494,
0.008765,
0.007318,
0.006109,
0.005108,
0.004263,
0.003556,
0.002967,
0.002475,
0.002064,
0.000833,
0.000336,
0.000135,
0.000054,
0.000022,
0.000009,
0.000004,
0.000001
],
"m_23": [
0.000851,
0.002133,
0.005304,
0.006357,
0.007615,
0.009117,
0.010909,
0.013044,
0.015586,
0.018609,
0.022196,
0.026449,
0.031478,
0.037414,
0.044401,
0.052601,
0.062193,
0.073369,
0.086332,
0.101291,
0.118455,
0.138019,
0.160154,
0.184993,
0.212609,
0.243005,
0.276092,
0.311685,
0.349489,
0.389109,
0.430058,
0.471785,
0.513695,
0.555191,
0.595705,
0.634724,
0.67182,
0.706662,
0.739018,
0.768757,
0.795835,
0.820286,
0.842202,
0.86172,
0.879006,
0.894243,
0.907621,
0.919327,
0.958731,
0.978157,
0.987801,
0.992741,
0.995392,
0.996894,
0.997796,
0.998751
],
"h1_37": [
0.978504,
0.948294,
0.880797,
0.860348,
0.837039,
0.810697,
0.781209,
0.748553,
0.712814,
0.674206,
0.63308,
0.589921,
0.54533,
0.5,
0.454671,
0.41008,
0.36692,
0.325794,
0.287186,
0.251448,
0.218791,
0.189304,
0.162961,
0.139653,
0.119203,
0.101396,
0.085988,
0.072733,
0.061383,
0.051706,
0.043484,
0.036519,
0.030634,
0.025672,
0.021496,
0.017986,
0.015041,
0.012572,
0.010504,
0.008773,
0.007325,
0.006115,
0.005103,
0.004259,
0.003553,
0.002964,
0.002473,
0.002062,
0.000832,
0.000335,
0.000135,
0.000054,
0.000022,
0.000009,
0.000004,
0.000001
],
"h1_23": [
0.978504,
0.948294,
0.880797,
0.860348,
0.837039,
0.810697,
0.781209,
0.748553,
0.712814,
0.674206,
0.63308,
0.589921,
0.54533,
0.5,
0.454671,
0.41008,
0.36692,
0.325794,
0.287186,
0.251448,
0.218791,
0.189304,
0.162961,
0.139653,
0.119203,
0.101396,
0.085988,
0.072733,
0.061383,
0.051706,
0.043484,
0.036519,
0.030634,
0.025672,
0.021496,
0.017986,
0.015041,
0.012572,
0.010504,
0.008773,
0.007325,
0.006115,
0.005103,
0.004259,
0.003553,
0.002964,
0.002473,
0.002062,
0.000832,
0.000335,
0.000135,
0.000054,
0.000022,
0.000009,
0.000004,
0.000001
]
}
SM1 FIT
false
SM2 FIT
false
SM3 FIT
false
SM4 FIT
false
SM5 FIT
false
Figures