139657_sjg_na.mod
Ion Channel Class: Na
SUFFIX
sjg_na
GMAX NAME
gnabar_sjg_na
G VALS
- gnabar_sjg_na: 0.07958
I VALS
{}
STATES
- m
- h
- p
GATES
- m: 0
- h: 1
- p: 1
ERROR FLAGS
- 4
RATES
true
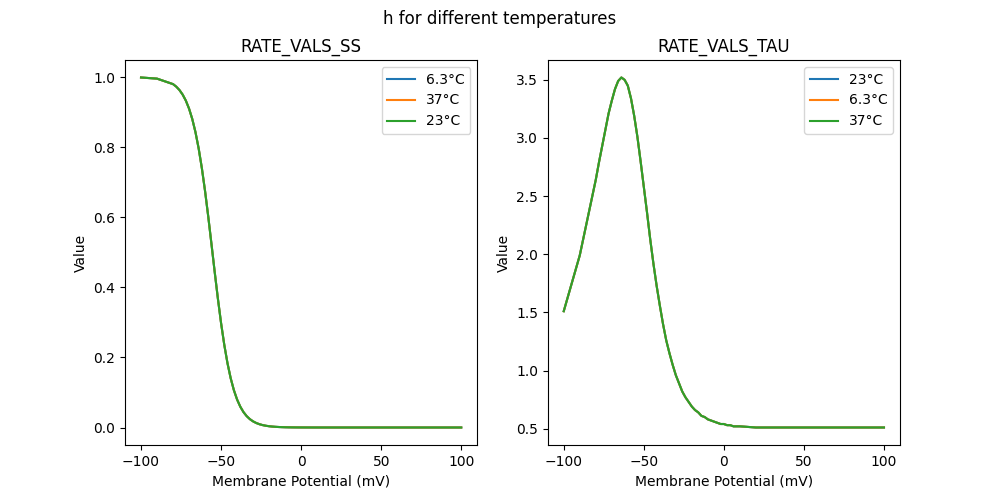
RATE VALS TAU
{
"h_23": [
1.51,
1.99,
2.64,
2.79,
2.93,
3.07,
3.21,
3.32,
3.42,
3.49,
3.52,
3.5,
3.45,
3.34,
3.19,
3.01,
2.8,
2.58,
2.36,
2.13,
1.92,
1.73,
1.56,
1.4,
1.26,
1.15,
1.05,
0.96,
0.89,
0.82,
0.77,
0.73,
0.69,
0.66,
0.64,
0.61,
0.6,
0.58,
0.57,
0.56,
0.55,
0.54,
0.54,
0.53,
0.53,
0.52,
0.52,
0.52,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51
],
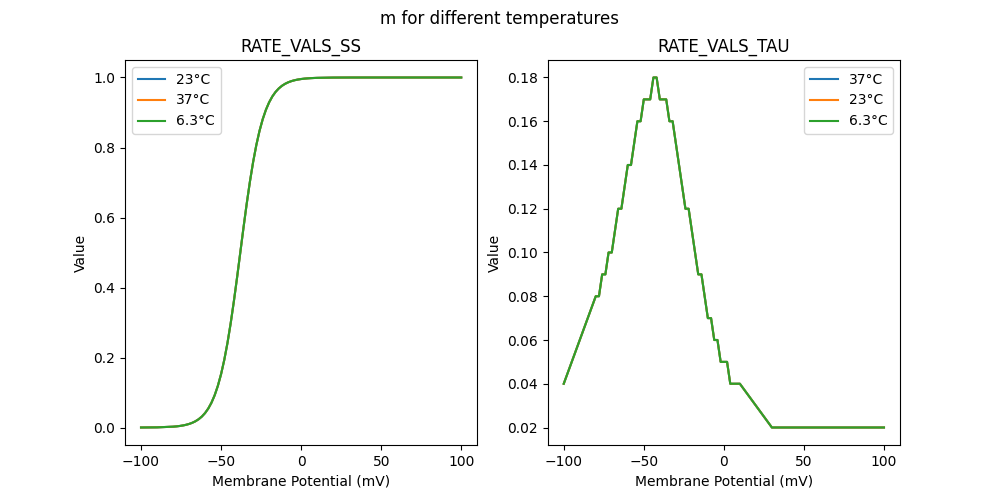
"m_37": [
0.04,
0.06,
0.08,
0.08,
0.09,
0.09,
0.1,
0.1,
0.11,
0.12,
0.12,
0.13,
0.14,
0.14,
0.15,
0.16,
0.16,
0.17,
0.17,
0.17,
0.18,
0.18,
0.17,
0.17,
0.17,
0.16,
0.16,
0.15,
0.14,
0.13,
0.12,
0.12,
0.11,
0.1,
0.09,
0.09,
0.08,
0.07,
0.07,
0.06,
0.06,
0.05,
0.05,
0.05,
0.04,
0.04,
0.04,
0.04,
0.03,
0.02,
0.02,
0.02,
0.02,
0.02,
0.02,
0.02
],
"p_6.3": [
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0
],
"p_37": [
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0
],
"p_23": [
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0
],
"m_23": [
0.04,
0.06,
0.08,
0.08,
0.09,
0.09,
0.1,
0.1,
0.11,
0.12,
0.12,
0.13,
0.14,
0.14,
0.15,
0.16,
0.16,
0.17,
0.17,
0.17,
0.18,
0.18,
0.17,
0.17,
0.17,
0.16,
0.16,
0.15,
0.14,
0.13,
0.12,
0.12,
0.11,
0.1,
0.09,
0.09,
0.08,
0.07,
0.07,
0.06,
0.06,
0.05,
0.05,
0.05,
0.04,
0.04,
0.04,
0.04,
0.03,
0.02,
0.02,
0.02,
0.02,
0.02,
0.02,
0.02
],
"m_6.3": [
0.04,
0.06,
0.08,
0.08,
0.09,
0.09,
0.1,
0.1,
0.11,
0.12,
0.12,
0.13,
0.14,
0.14,
0.15,
0.16,
0.16,
0.17,
0.17,
0.17,
0.18,
0.18,
0.17,
0.17,
0.17,
0.16,
0.16,
0.15,
0.14,
0.13,
0.12,
0.12,
0.11,
0.1,
0.09,
0.09,
0.08,
0.07,
0.07,
0.06,
0.06,
0.05,
0.05,
0.05,
0.04,
0.04,
0.04,
0.04,
0.03,
0.02,
0.02,
0.02,
0.02,
0.02,
0.02,
0.02
],
"h_6.3": [
1.51,
1.99,
2.64,
2.79,
2.93,
3.07,
3.21,
3.32,
3.42,
3.49,
3.52,
3.5,
3.45,
3.34,
3.19,
3.01,
2.8,
2.58,
2.36,
2.13,
1.92,
1.73,
1.56,
1.4,
1.26,
1.15,
1.05,
0.96,
0.89,
0.82,
0.77,
0.73,
0.69,
0.66,
0.64,
0.61,
0.6,
0.58,
0.57,
0.56,
0.55,
0.54,
0.54,
0.53,
0.53,
0.52,
0.52,
0.52,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51
],
"h_37": [
1.51,
1.99,
2.64,
2.79,
2.93,
3.07,
3.21,
3.32,
3.42,
3.49,
3.52,
3.5,
3.45,
3.34,
3.19,
3.01,
2.8,
2.58,
2.36,
2.13,
1.92,
1.73,
1.56,
1.4,
1.26,
1.15,
1.05,
0.96,
0.89,
0.82,
0.77,
0.73,
0.69,
0.66,
0.64,
0.61,
0.6,
0.58,
0.57,
0.56,
0.55,
0.54,
0.54,
0.53,
0.53,
0.52,
0.52,
0.52,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51,
0.51
]
}
RATE VALS SS
{
"p_23": [
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0
],
"m_23": [
0.000142,
0.000594,
0.002473,
0.003288,
0.00437,
0.005807,
0.007713,
0.010237,
0.013577,
0.017986,
0.023793,
0.031414,
0.041374,
0.054313,
0.071,
0.092313,
0.119203,
0.152608,
0.193321,
0.241796,
0.297937,
0.360908,
0.429055,
0.500002,
0.570948,
0.639094,
0.702065,
0.758205,
0.806679,
0.847392,
0.880797,
0.907687,
0.929,
0.945687,
0.958626,
0.968585,
0.976207,
0.982014,
0.986423,
0.989763,
0.992287,
0.994193,
0.99563,
0.996712,
0.997527,
0.998141,
0.998602,
0.998949,
0.999748,
0.99994,
0.999986,
0.999997,
0.999999,
1,
1,
1
],
"h_6.3": [
0.999158,
0.995898,
0.980252,
0.973072,
0.963379,
0.950376,
0.933075,
0.910312,
0.880797,
0.84324,
0.796584,
0.740318,
0.67484,
0.601735,
0.523792,
0.444672,
0.368262,
0.297937,
0.236024,
0.183612,
0.140695,
0.106501,
0.079845,
0.059417,
0.043966,
0.032395,
0.023793,
0.017434,
0.012752,
0.009316,
0.006799,
0.004959,
0.003615,
0.002634,
0.001919,
0.001398,
0.001018,
0.000741,
0.00054,
0.000393,
0.000286,
0.000208,
0.000152,
0.00011,
0.00008,
0.000059,
0.000043,
0.000031,
0.000006,
0.000001,
0,
0,
0,
0,
0,
0
],
"m_37": [
0.000142,
0.000594,
0.002473,
0.003288,
0.00437,
0.005807,
0.007713,
0.010237,
0.013577,
0.017986,
0.023793,
0.031414,
0.041374,
0.054313,
0.071,
0.092313,
0.119203,
0.152608,
0.193321,
0.241796,
0.297937,
0.360908,
0.429055,
0.500002,
0.570948,
0.639094,
0.702065,
0.758205,
0.806679,
0.847392,
0.880797,
0.907687,
0.929,
0.945687,
0.958626,
0.968585,
0.976207,
0.982014,
0.986423,
0.989763,
0.992287,
0.994193,
0.99563,
0.996712,
0.997527,
0.998141,
0.998602,
0.998949,
0.999748,
0.99994,
0.999986,
0.999997,
0.999999,
1,
1,
1
],
"p_6.3": [
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0
],
"p_37": [
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0
],
"m_6.3": [
0.000142,
0.000594,
0.002473,
0.003288,
0.00437,
0.005807,
0.007713,
0.010237,
0.013577,
0.017986,
0.023793,
0.031414,
0.041374,
0.054313,
0.071,
0.092313,
0.119203,
0.152608,
0.193321,
0.241796,
0.297937,
0.360908,
0.429055,
0.500002,
0.570948,
0.639094,
0.702065,
0.758205,
0.806679,
0.847392,
0.880797,
0.907687,
0.929,
0.945687,
0.958626,
0.968585,
0.976207,
0.982014,
0.986423,
0.989763,
0.992287,
0.994193,
0.99563,
0.996712,
0.997527,
0.998141,
0.998602,
0.998949,
0.999748,
0.99994,
0.999986,
0.999997,
0.999999,
1,
1,
1
],
"h_37": [
0.999158,
0.995898,
0.980252,
0.973072,
0.963379,
0.950376,
0.933075,
0.910312,
0.880797,
0.84324,
0.796584,
0.740318,
0.67484,
0.601735,
0.523792,
0.444672,
0.368262,
0.297937,
0.236024,
0.183612,
0.140695,
0.106501,
0.079845,
0.059417,
0.043966,
0.032395,
0.023793,
0.017434,
0.012752,
0.009316,
0.006799,
0.004959,
0.003615,
0.002634,
0.001919,
0.001398,
0.001018,
0.000741,
0.00054,
0.000393,
0.000286,
0.000208,
0.000152,
0.00011,
0.00008,
0.000059,
0.000043,
0.000031,
0.000006,
0.000001,
0,
0,
0,
0,
0,
0
],
"h_23": [
0.999158,
0.995898,
0.980252,
0.973072,
0.963379,
0.950376,
0.933075,
0.910312,
0.880797,
0.84324,
0.796584,
0.740318,
0.67484,
0.601735,
0.523792,
0.444672,
0.368262,
0.297937,
0.236024,
0.183612,
0.140695,
0.106501,
0.079845,
0.059417,
0.043966,
0.032395,
0.023793,
0.017434,
0.012752,
0.009316,
0.006799,
0.004959,
0.003615,
0.002634,
0.001919,
0.001398,
0.001018,
0.000741,
0.00054,
0.000393,
0.000286,
0.000208,
0.000152,
0.00011,
0.00008,
0.000059,
0.000043,
0.000031,
0.000006,
0.000001,
0,
0,
0,
0,
0,
0
]
}
SM1 FIT
false
SM2 FIT
false
SM3 FIT
false
SM4 FIT
false
SM5 FIT
false
Figures