151460_AXNODE75.mod
Ion Channel Class: Na
SUFFIX
axnode75
GMAX NAME
gnabar_axnode75
G VALS
- gkbar_axnode75: 0
- gnabar_axnode75: 3
- gnapbar_axnode75: 0
I VALS
- 37: 7.678181721895223
- 6.3: 80.77033047650552
STATES
- mp
- m
- h
- s
GATES
- mp: 3
- m: 6
- h: 1
- s: 1
ERROR FLAGS
- 19
RATES
true
RATE VALS TAU
{
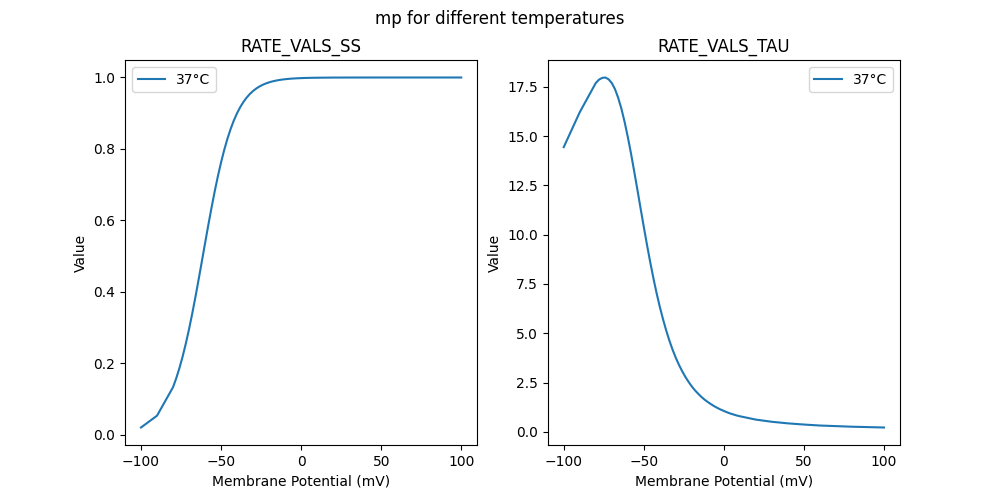
"mp_37": [
14.44,
16.21,
17.69,
17.86,
17.95,
17.97,
17.88,
17.69,
17.38,
16.94,
16.39,
15.72,
14.96,
14.12,
13.21,
12.28,
11.33,
10.39,
9.48,
8.61,
7.79,
7.03,
6.34,
5.71,
5.14,
4.62,
4.16,
3.75,
3.39,
3.07,
2.78,
2.53,
2.3,
2.1,
1.93,
1.77,
1.63,
1.51,
1.4,
1.3,
1.21,
1.13,
1.06,
0.99,
0.93,
0.88,
0.83,
0.79,
0.62,
0.51,
0.43,
0.37,
0.32,
0.29,
0.26,
0.22
],
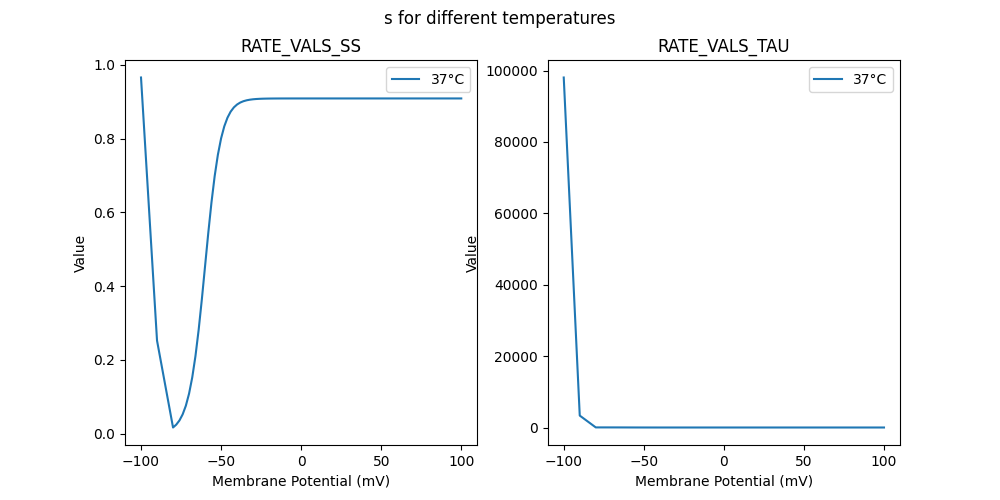
"s_37": [
98039.550015,
3340.15,
29.58,
29.18,
28.81,
28.32,
27.62,
26.64,
25.32,
23.6,
21.46,
18.99,
16.31,
13.63,
11.15,
9.01,
7.29,
5.96,
4.98,
4.28,
3.78,
3.44,
3.21,
3.05,
2.94,
2.87,
2.82,
2.79,
2.77,
2.75,
2.74,
2.73,
2.73,
2.73,
2.72,
2.72,
2.72,
2.72,
2.72,
2.72,
2.72,
2.72,
2.72,
2.72,
2.72,
2.72,
2.72,
2.72,
2.72,
2.72,
2.72,
2.72,
2.72,
2.72,
2.72,
2.72
],
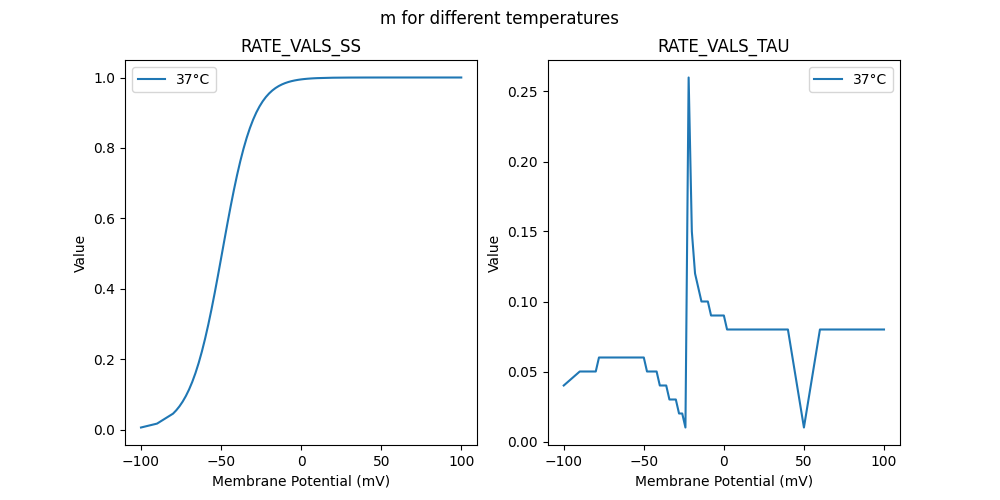
"m_37": [
0.04,
0.05,
0.05,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.06,
0.05,
0.05,
0.05,
0.05,
0.04,
0.04,
0.04,
0.03,
0.03,
0.03,
0.02,
0.02,
0.01,
0.26,
0.15,
0.12,
0.11,
0.1,
0.1,
0.1,
0.09,
0.09,
0.09,
0.09,
0.09,
0.08,
0.08,
0.08,
0.08,
0.08,
0.08,
0.08,
0.08,
0.01,
0.08,
0.08,
0.08,
0.08
],
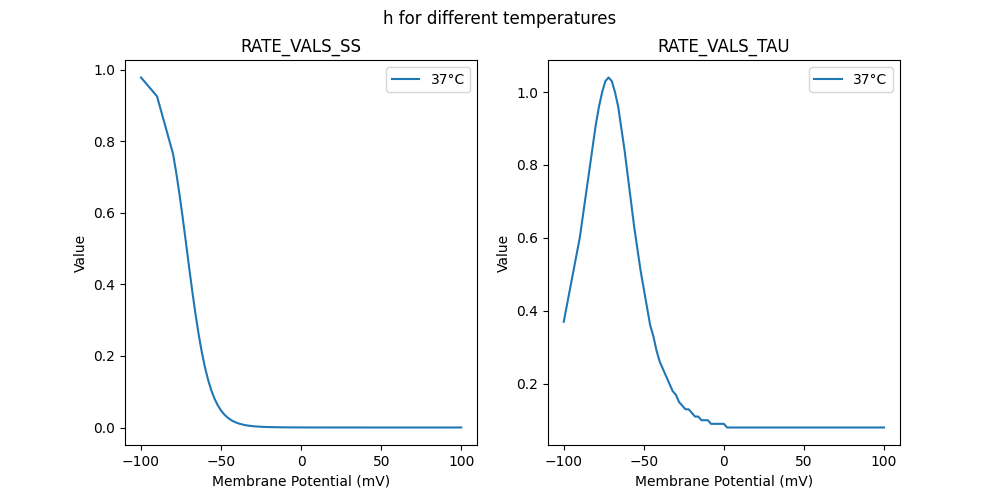
"h_37": [
0.37,
0.6,
0.91,
0.96,
1,
1.03,
1.04,
1.03,
1,
0.96,
0.9,
0.84,
0.77,
0.7,
0.63,
0.57,
0.51,
0.46,
0.41,
0.36,
0.33,
0.29,
0.26,
0.24,
0.22,
0.2,
0.18,
0.17,
0.15,
0.14,
0.13,
0.13,
0.12,
0.11,
0.11,
0.1,
0.1,
0.1,
0.09,
0.09,
0.09,
0.09,
0.09,
0.08,
0.08,
0.08,
0.08,
0.08,
0.08,
0.08,
0.08,
0.08,
0.08,
0.08,
0.08,
0.08
]
}
RATE VALS SS
{
"mp_37": [
0.020543,
0.053651,
0.13337,
0.158276,
0.186868,
0.219325,
0.255701,
0.295887,
0.339577,
0.38626,
0.435217,
0.485563,
0.536298,
0.586389,
0.634843,
0.680801,
0.723576,
0.762694,
0.797891,
0.829097,
0.856403,
0.880021,
0.900243,
0.917404,
0.93186,
0.943958,
0.954026,
0.962368,
0.969252,
0.974913,
0.979557,
0.983358,
0.986463,
0.988995,
0.991057,
0.992735,
0.994099,
0.995208,
0.996109,
0.99684,
0.997433,
0.997915,
0.998306,
0.998623,
0.998881,
0.99909,
0.99926,
0.999398,
0.999785,
0.999923,
0.999972,
0.99999,
0.999996,
0.999999,
1,
1
],
"m_37": [
0.006762,
0.01779,
0.046079,
0.055534,
0.066804,
0.080181,
0.095979,
0.114525,
0.136143,
0.161136,
0.189755,
0.222166,
0.258415,
0.298389,
0.34179,
0.388117,
0.436677,
0.486611,
0.536952,
0.586695,
0.634877,
0.680649,
0.72333,
0.762436,
0.79769,
0.829004,
0.85645,
0.880219,
0.900591,
0.917888,
0.932459,
0.944646,
0.954779,
0.96316,
0.97006,
0.975718,
0.980343,
0.984113,
0.987177,
0.989662,
0.991674,
0.9933,
0.994612,
0.99567,
0.996522,
0.997208,
0.997759,
0.998202,
0.999403,
0.999802,
0.999934,
0.999978,
0.999993,
0.999998,
0.999999,
1
],
"s_37": [
0.965919,
0.252277,
0.016426,
0.024152,
0.035539,
0.052013,
0.075471,
0.108175,
0.152459,
0.210119,
0.281473,
0.364428,
0.454139,
0.543882,
0.62692,
0.698392,
0.756175,
0.800572,
0.833369,
0.856899,
0.87343,
0.884873,
0.892712,
0.898046,
0.901657,
0.904094,
0.905735,
0.906839,
0.90758,
0.908078,
0.908411,
0.908635,
0.908785,
0.908886,
0.908954,
0.908999,
0.909029,
0.90905,
0.909063,
0.909072,
0.909078,
0.909083,
0.909085,
0.909087,
0.909088,
0.909089,
0.90909,
0.90909,
0.909091,
0.909091,
0.909091,
0.909091,
0.909091,
0.909091,
0.909091,
0.909091
],
"h_37": [
0.978426,
0.925855,
0.765047,
0.712263,
0.652668,
0.587553,
0.518935,
0.449352,
0.381507,
0.317858,
0.260281,
0.209894,
0.167068,
0.131561,
0.102726,
0.079696,
0.061545,
0.047384,
0.03642,
0.027979,
0.021505,
0.016552,
0.012766,
0.009872,
0.00766,
0.005965,
0.004665,
0.003664,
0.002891,
0.002292,
0.001826,
0.001462,
0.001176,
0.000951,
0.000772,
0.000629,
0.000515,
0.000423,
0.000349,
0.000289,
0.000239,
0.000199,
0.000166,
0.000138,
0.000116,
0.000097,
0.000081,
0.000068,
0.000029,
0.000012,
0.000005,
0.000002,
0.000001,
0,
0,
0
]
}
SM1 FIT
false
SM2 FIT
false
SM3 FIT
false
SM4 FIT
false
SM5 FIT
false
Figures