155796_nav19.mod
Ion Channel Class: Na
SUFFIX
nap
GMAX NAME
gbar_nap
G VALS
- gbar_nap: 0.00001
I VALS
- 37: 0.028402734214733008
- 6.3: 0.028402734214733008
STATES
- m
GATES
- m: 2
ERROR FLAGS
- 8
RATES
true
Q10 TAU
1
Q10 G
1
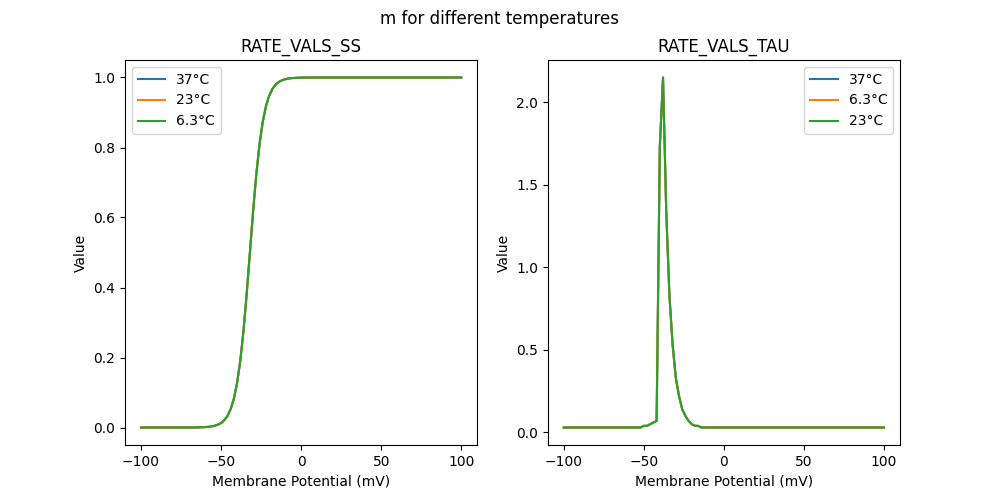
RATE VALS TAU
{
"m_37": [
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.04,
0.04,
0.05,
0.06,
0.07,
1.73,
2.15,
1.34,
0.83,
0.53,
0.33,
0.22,
0.14,
0.1,
0.07,
0.05,
0.04,
0.04,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03
],
"m_6.3": [
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.04,
0.04,
0.05,
0.06,
0.07,
1.73,
2.15,
1.34,
0.83,
0.53,
0.33,
0.22,
0.14,
0.1,
0.07,
0.05,
0.04,
0.04,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03
],
"m_23": [
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.04,
0.04,
0.05,
0.06,
0.07,
1.73,
2.15,
1.34,
0.83,
0.53,
0.33,
0.22,
0.14,
0.1,
0.07,
0.05,
0.04,
0.04,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03,
0.03
]
}
RATE VALS SS
{
"m_37": [
0,
0.000001,
0.000009,
0.000015,
0.000025,
0.00004,
0.000065,
0.000106,
0.000171,
0.000277,
0.000448,
0.000725,
0.001174,
0.001899,
0.003071,
0.004963,
0.008011,
0.012908,
0.020735,
0.033147,
0.052592,
0.082469,
0.127041,
0.190693,
0.276145,
0.381819,
0.499997,
0.618175,
0.723851,
0.809304,
0.872958,
0.917531,
0.947408,
0.966853,
0.979266,
0.987092,
0.991989,
0.995037,
0.996929,
0.998101,
0.998827,
0.999275,
0.999552,
0.999723,
0.999829,
0.999894,
0.999935,
0.99996,
0.999996,
1,
1,
1,
1,
1,
1,
1
],
"m_23": [
0,
0.000001,
0.000009,
0.000015,
0.000025,
0.00004,
0.000065,
0.000106,
0.000171,
0.000277,
0.000448,
0.000725,
0.001174,
0.001899,
0.003071,
0.004963,
0.008011,
0.012908,
0.020735,
0.033147,
0.052592,
0.082469,
0.127041,
0.190693,
0.276145,
0.381819,
0.499997,
0.618175,
0.723851,
0.809304,
0.872958,
0.917531,
0.947408,
0.966853,
0.979266,
0.987092,
0.991989,
0.995037,
0.996929,
0.998101,
0.998827,
0.999275,
0.999552,
0.999723,
0.999829,
0.999894,
0.999935,
0.99996,
0.999996,
1,
1,
1,
1,
1,
1,
1
],
"m_6.3": [
0,
0.000001,
0.000009,
0.000015,
0.000025,
0.00004,
0.000065,
0.000106,
0.000171,
0.000277,
0.000448,
0.000725,
0.001174,
0.001899,
0.003071,
0.004963,
0.008011,
0.012908,
0.020735,
0.033147,
0.052592,
0.082469,
0.127041,
0.190693,
0.276145,
0.381819,
0.499997,
0.618175,
0.723851,
0.809304,
0.872958,
0.917531,
0.947408,
0.966853,
0.979266,
0.987092,
0.991989,
0.995037,
0.996929,
0.998101,
0.998827,
0.999275,
0.999552,
0.999723,
0.999829,
0.999894,
0.999935,
0.99996,
0.999996,
1,
1,
1,
1,
1,
1,
1
]
}
SM1 FIT
false
SM2 FIT
false
SM3 FIT
false
SM4 FIT
false
SM5 FIT
false
Figures