182797_Nadend.mod
Ion Channel Class: Na
SUFFIX
Nadend
GMAX NAME
null
G VALS
{}
I VALS
{}
STATES
- m
- h
GATES
- m: 3
- h: 1
ERROR FLAGS
- 5
RATES
true
RATE VALS TAU
{
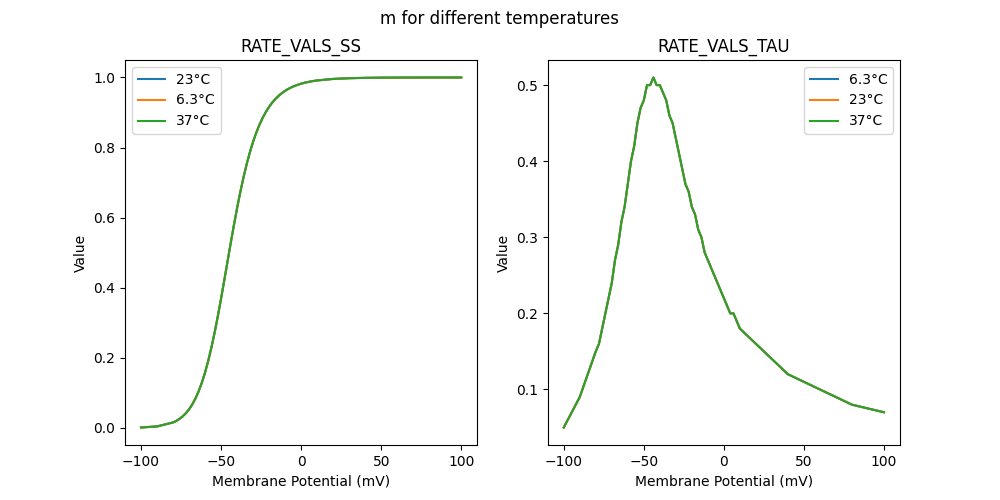
"m_6.3": [
0.05,
0.09,
0.15,
0.16,
0.18,
0.2,
0.22,
0.24,
0.27,
0.29,
0.32,
0.34,
0.37,
0.4,
0.42,
0.45,
0.47,
0.48,
0.5,
0.5,
0.51,
0.5,
0.5,
0.49,
0.48,
0.46,
0.45,
0.43,
0.41,
0.39,
0.37,
0.36,
0.34,
0.33,
0.31,
0.3,
0.28,
0.27,
0.26,
0.25,
0.24,
0.23,
0.22,
0.21,
0.2,
0.2,
0.19,
0.18,
0.16,
0.14,
0.12,
0.11,
0.1,
0.09,
0.08,
0.07
],
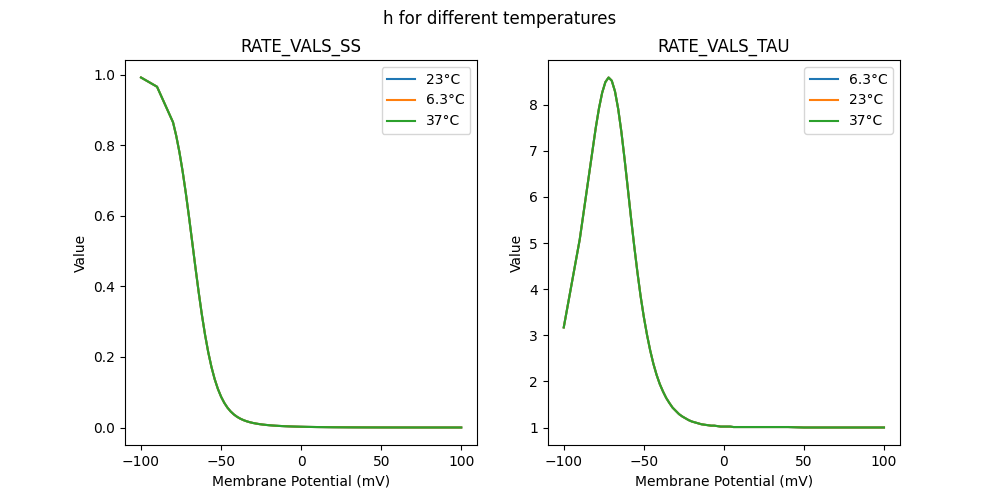
"h_6.3": [
3.17,
5.08,
7.5,
7.92,
8.26,
8.49,
8.59,
8.52,
8.29,
7.91,
7.41,
6.82,
6.19,
5.55,
4.94,
4.37,
3.85,
3.4,
3.01,
2.67,
2.38,
2.14,
1.94,
1.78,
1.64,
1.53,
1.43,
1.36,
1.29,
1.24,
1.2,
1.16,
1.13,
1.11,
1.09,
1.07,
1.06,
1.05,
1.04,
1.04,
1.03,
1.02,
1.02,
1.02,
1.02,
1.01,
1.01,
1.01,
1.01,
1.01,
1.01,
1,
1,
1,
1,
1
],
"m_23": [
0.05,
0.09,
0.15,
0.16,
0.18,
0.2,
0.22,
0.24,
0.27,
0.29,
0.32,
0.34,
0.37,
0.4,
0.42,
0.45,
0.47,
0.48,
0.5,
0.5,
0.51,
0.5,
0.5,
0.49,
0.48,
0.46,
0.45,
0.43,
0.41,
0.39,
0.37,
0.36,
0.34,
0.33,
0.31,
0.3,
0.28,
0.27,
0.26,
0.25,
0.24,
0.23,
0.22,
0.21,
0.2,
0.2,
0.19,
0.18,
0.16,
0.14,
0.12,
0.11,
0.1,
0.09,
0.08,
0.07
],
"h_23": [
3.17,
5.08,
7.5,
7.92,
8.26,
8.49,
8.59,
8.52,
8.29,
7.91,
7.41,
6.82,
6.19,
5.55,
4.94,
4.37,
3.85,
3.4,
3.01,
2.67,
2.38,
2.14,
1.94,
1.78,
1.64,
1.53,
1.43,
1.36,
1.29,
1.24,
1.2,
1.16,
1.13,
1.11,
1.09,
1.07,
1.06,
1.05,
1.04,
1.04,
1.03,
1.02,
1.02,
1.02,
1.02,
1.01,
1.01,
1.01,
1.01,
1.01,
1.01,
1,
1,
1,
1,
1
],
"h_37": [
3.17,
5.08,
7.5,
7.92,
8.26,
8.49,
8.59,
8.52,
8.29,
7.91,
7.41,
6.82,
6.19,
5.55,
4.94,
4.37,
3.85,
3.4,
3.01,
2.67,
2.38,
2.14,
1.94,
1.78,
1.64,
1.53,
1.43,
1.36,
1.29,
1.24,
1.2,
1.16,
1.13,
1.11,
1.09,
1.07,
1.06,
1.05,
1.04,
1.04,
1.03,
1.02,
1.02,
1.02,
1.02,
1.01,
1.01,
1.01,
1.01,
1.01,
1.01,
1,
1,
1,
1,
1
],
"m_37": [
0.05,
0.09,
0.15,
0.16,
0.18,
0.2,
0.22,
0.24,
0.27,
0.29,
0.32,
0.34,
0.37,
0.4,
0.42,
0.45,
0.47,
0.48,
0.5,
0.5,
0.51,
0.5,
0.5,
0.49,
0.48,
0.46,
0.45,
0.43,
0.41,
0.39,
0.37,
0.36,
0.34,
0.33,
0.31,
0.3,
0.28,
0.27,
0.26,
0.25,
0.24,
0.23,
0.22,
0.21,
0.2,
0.2,
0.19,
0.18,
0.16,
0.14,
0.12,
0.11,
0.1,
0.09,
0.08,
0.07
]
}
RATE VALS SS
{
"m_23": [
0.001065,
0.004143,
0.015392,
0.019855,
0.025528,
0.032698,
0.041703,
0.052933,
0.066818,
0.083823,
0.104416,
0.129038,
0.158052,
0.191688,
0.229978,
0.272707,
0.319378,
0.369216,
0.421207,
0.474178,
0.526907,
0.57823,
0.627142,
0.672859,
0.714852,
0.752834,
0.786736,
0.816659,
0.842824,
0.865531,
0.885119,
0.901936,
0.916324,
0.928602,
0.939061,
0.94796,
0.955529,
0.961965,
0.967438,
0.972096,
0.976062,
0.979442,
0.982326,
0.984789,
0.986894,
0.988697,
0.990241,
0.991566,
0.995877,
0.997944,
0.998957,
0.999464,
0.999722,
0.999854,
0.999923,
0.999978
],
"h_23": [
0.99218,
0.96602,
0.865166,
0.826761,
0.780347,
0.725828,
0.663891,
0.596119,
0.524921,
0.453232,
0.384042,
0.319911,
0.262632,
0.21311,
0.171454,
0.137188,
0.109489,
0.087384,
0.069905,
0.056159,
0.04538,
0.036929,
0.030292,
0.02506,
0.020913,
0.017606,
0.014947,
0.012793,
0.011031,
0.009576,
0.008365,
0.007346,
0.006481,
0.005742,
0.005105,
0.004552,
0.004069,
0.003645,
0.003271,
0.00294,
0.002645,
0.002383,
0.002148,
0.001938,
0.001749,
0.001579,
0.001427,
0.001289,
0.000779,
0.000472,
0.000286,
0.000174,
0.000105,
0.000064,
0.000039,
0.000014
],
"m_6.3": [
0.001065,
0.004143,
0.015392,
0.019855,
0.025528,
0.032698,
0.041703,
0.052933,
0.066818,
0.083823,
0.104416,
0.129038,
0.158052,
0.191688,
0.229978,
0.272707,
0.319378,
0.369216,
0.421207,
0.474178,
0.526907,
0.57823,
0.627142,
0.672859,
0.714852,
0.752834,
0.786736,
0.816659,
0.842824,
0.865531,
0.885119,
0.901936,
0.916324,
0.928602,
0.939061,
0.94796,
0.955529,
0.961965,
0.967438,
0.972096,
0.976062,
0.979442,
0.982326,
0.984789,
0.986894,
0.988697,
0.990241,
0.991566,
0.995877,
0.997944,
0.998957,
0.999464,
0.999722,
0.999854,
0.999923,
0.999978
],
"h_6.3": [
0.99218,
0.96602,
0.865166,
0.826761,
0.780347,
0.725828,
0.663891,
0.596119,
0.524921,
0.453232,
0.384042,
0.319911,
0.262632,
0.21311,
0.171454,
0.137188,
0.109489,
0.087384,
0.069905,
0.056159,
0.04538,
0.036929,
0.030292,
0.02506,
0.020913,
0.017606,
0.014947,
0.012793,
0.011031,
0.009576,
0.008365,
0.007346,
0.006481,
0.005742,
0.005105,
0.004552,
0.004069,
0.003645,
0.003271,
0.00294,
0.002645,
0.002383,
0.002148,
0.001938,
0.001749,
0.001579,
0.001427,
0.001289,
0.000779,
0.000472,
0.000286,
0.000174,
0.000105,
0.000064,
0.000039,
0.000014
],
"m_37": [
0.001065,
0.004143,
0.015392,
0.019855,
0.025528,
0.032698,
0.041703,
0.052933,
0.066818,
0.083823,
0.104416,
0.129038,
0.158052,
0.191688,
0.229978,
0.272707,
0.319378,
0.369216,
0.421207,
0.474178,
0.526907,
0.57823,
0.627142,
0.672859,
0.714852,
0.752834,
0.786736,
0.816659,
0.842824,
0.865531,
0.885119,
0.901936,
0.916324,
0.928602,
0.939061,
0.94796,
0.955529,
0.961965,
0.967438,
0.972096,
0.976062,
0.979442,
0.982326,
0.984789,
0.986894,
0.988697,
0.990241,
0.991566,
0.995877,
0.997944,
0.998957,
0.999464,
0.999722,
0.999854,
0.999923,
0.999978
],
"h_37": [
0.99218,
0.96602,
0.865166,
0.826761,
0.780347,
0.725828,
0.663891,
0.596119,
0.524921,
0.453232,
0.384042,
0.319911,
0.262632,
0.21311,
0.171454,
0.137188,
0.109489,
0.087384,
0.069905,
0.056159,
0.04538,
0.036929,
0.030292,
0.02506,
0.020913,
0.017606,
0.014947,
0.012793,
0.011031,
0.009576,
0.008365,
0.007346,
0.006481,
0.005742,
0.005105,
0.004552,
0.004069,
0.003645,
0.003271,
0.00294,
0.002645,
0.002383,
0.002148,
0.001938,
0.001749,
0.001579,
0.001427,
0.001289,
0.000779,
0.000472,
0.000286,
0.000174,
0.000105,
0.000064,
0.000039,
0.000014
]
}
SM1 FIT
false
SM2 FIT
false
SM3 FIT
false
SM4 FIT
false
SM5 FIT
false
Figures