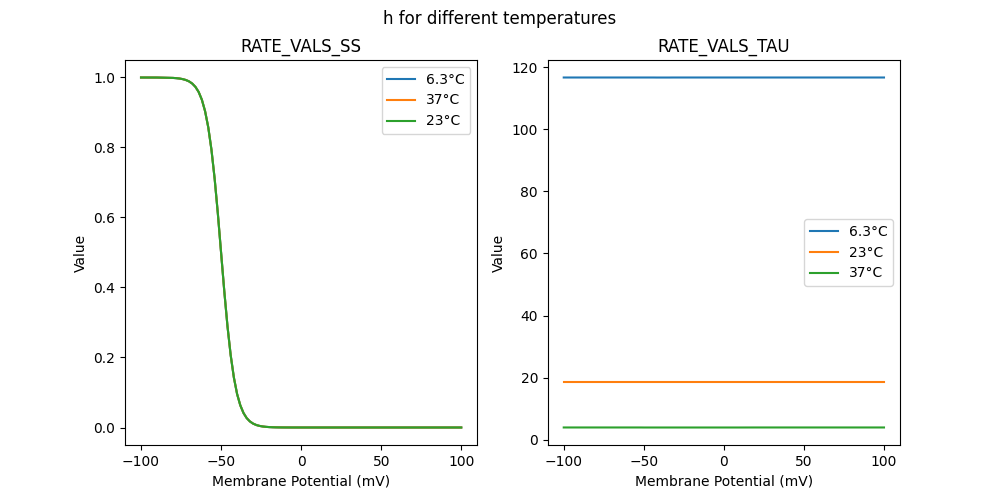
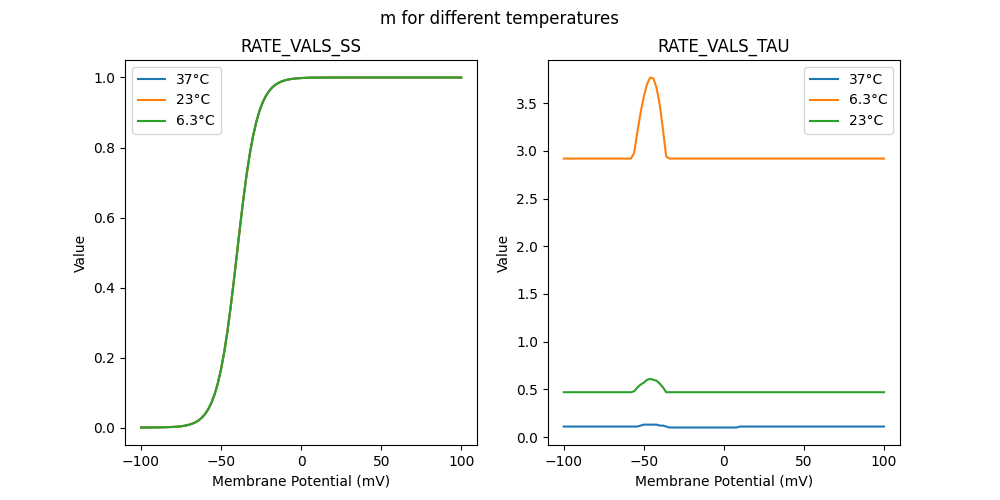
52034_nap.mod
Ion Channel Class: Na
SUFFIX
nap
GMAX NAME
gmax_nap
G VALS
- gmax_nap: 0.1
I VALS
{}
STATES
- m
- h
GATES
- m: 0
- h: 2
ERROR FLAGS
- 4
RATES
true
RATE VALS TAU
{
"m_37": [
0.11,
0.11,
0.11,
0.11,
0.11,
0.11,
0.11,
0.11,
0.11,
0.11,
0.11,
0.11,
0.11,
0.11,
0.11,
0.11,
0.12,
0.13,
0.13,
0.13,
0.13,
0.13,
0.12,
0.12,
0.11,
0.1,
0.1,
0.1,
0.1,
0.1,
0.1,
0.1,
0.1,
0.1,
0.1,
0.1,
0.1,
0.1,
0.1,
0.1,
0.1,
0.1,
0.1,
0.1,
0.1,
0.1,
0.1,
0.11,
0.11,
0.11,
0.11,
0.11,
0.11,
0.11,
0.11,
0.11
],
"h_6.3": [
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.63,
116.63,
116.63,
116.63,
116.63,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64,
116.64
],
"h_23": [
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63,
18.63
],
"m_6.3": [
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.98,
3.2,
3.4,
3.57,
3.7,
3.77,
3.76,
3.66,
3.48,
3.22,
2.94,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92,
2.92
],
"h_37": [
4.01,
4.01,
4.01,
4.01,
4.01,
4.01,
4.01,
4.01,
4.01,
4.01,
4.01,
4.01,
4.01,
4.01,
4.01,
4.01,
4.01,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4.01,
4.01,
4.01,
4.01,
4.01,
4.01,
4.01,
4.01
],
"m_23": [
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.48,
0.52,
0.55,
0.57,
0.6,
0.61,
0.6,
0.59,
0.56,
0.52,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47,
0.47
]
}
RATE VALS SS
{
"h_6.3": [
0.999884,
0.999771,
0.998707,
0.998033,
0.996989,
0.995334,
0.992765,
0.988803,
0.982573,
0.973018,
0.958536,
0.936418,
0.903907,
0.857578,
0.79326,
0.7102,
0.610341,
0.499941,
0.389725,
0.289425,
0.206635,
0.142589,
0.095765,
0.063482,
0.041473,
0.0268,
0.017325,
0.011127,
0.007109,
0.004564,
0.002917,
0.001858,
0.001191,
0.00076,
0.000484,
0.00031,
0.000198,
0.000126,
0.000081,
0.000051,
0.000033,
0.000021,
0.000013,
0.000009,
0.000005,
0.000003,
0.000002,
0.000001,
0,
0,
0,
0,
0,
0,
0,
0
],
"h_37": [
0.999986,
0.999873,
0.998809,
0.998135,
0.997091,
0.995436,
0.992867,
0.988905,
0.982674,
0.973118,
0.958634,
0.936514,
0.903999,
0.857666,
0.793341,
0.710273,
0.610404,
0.499992,
0.389765,
0.289455,
0.206656,
0.142604,
0.095775,
0.063489,
0.041477,
0.026802,
0.017327,
0.011128,
0.00711,
0.004564,
0.002918,
0.001858,
0.001191,
0.00076,
0.000484,
0.00031,
0.000198,
0.000126,
0.000081,
0.000051,
0.000033,
0.000021,
0.000013,
0.000009,
0.000005,
0.000003,
0.000002,
0.000001,
0,
0,
0,
0,
0,
0,
0,
0
],
"m_37": [
0.000064,
0.000321,
0.001603,
0.00221,
0.003042,
0.004198,
0.005783,
0.007948,
0.010949,
0.015043,
0.020603,
0.028245,
0.038553,
0.052346,
0.070854,
0.09519,
0.126668,
0.166826,
0.216447,
0.275806,
0.344584,
0.4204,
0.500005,
0.579912,
0.655502,
0.724199,
0.783512,
0.833117,
0.873332,
0.904755,
0.9291,
0.947654,
0.961446,
0.971755,
0.979396,
0.984957,
0.989051,
0.992052,
0.994217,
0.995802,
0.996958,
0.99779,
0.998397,
0.998839,
0.999157,
0.999389,
0.999558,
0.999679,
0.999936,
0.999987,
0.999997,
0.999999,
0.999999,
0.999999,
0.999999,
0.999999
],
"m_23": [
0.000064,
0.000321,
0.001603,
0.00221,
0.003042,
0.004198,
0.005783,
0.007948,
0.010949,
0.015043,
0.020603,
0.028245,
0.038553,
0.052346,
0.070852,
0.095188,
0.126668,
0.166824,
0.216446,
0.275806,
0.344585,
0.420403,
0.500005,
0.579919,
0.655504,
0.724199,
0.783512,
0.833117,
0.873332,
0.904755,
0.9291,
0.947654,
0.961446,
0.971755,
0.979396,
0.984957,
0.989051,
0.992052,
0.994217,
0.995802,
0.996958,
0.99779,
0.998397,
0.998839,
0.999157,
0.999389,
0.999558,
0.999679,
0.999936,
0.999987,
0.999997,
0.999999,
0.999999,
0.999999,
0.999999,
0.999999
],
"h_23": [
0.999974,
0.999861,
0.998797,
0.998123,
0.997079,
0.995424,
0.992855,
0.988893,
0.982662,
0.973106,
0.958623,
0.936503,
0.903988,
0.857656,
0.793331,
0.710264,
0.610396,
0.499986,
0.389761,
0.289451,
0.206653,
0.142602,
0.095774,
0.063488,
0.041477,
0.026802,
0.017327,
0.011128,
0.00711,
0.004564,
0.002918,
0.001858,
0.001191,
0.00076,
0.000484,
0.00031,
0.000198,
0.000126,
0.000081,
0.000051,
0.000033,
0.000021,
0.000013,
0.000009,
0.000005,
0.000003,
0.000002,
0.000001,
0,
0,
0,
0,
0,
0,
0,
0
],
"m_6.3": [
0.000064,
0.000321,
0.001603,
0.00221,
0.003042,
0.004198,
0.005783,
0.007948,
0.010949,
0.015043,
0.020603,
0.028245,
0.038553,
0.052346,
0.070851,
0.095188,
0.126668,
0.166823,
0.216446,
0.275806,
0.344585,
0.420404,
0.500005,
0.579921,
0.655504,
0.724199,
0.783512,
0.833117,
0.873332,
0.904755,
0.9291,
0.947654,
0.961446,
0.971755,
0.979396,
0.984957,
0.989051,
0.992052,
0.994217,
0.995802,
0.996958,
0.99779,
0.998397,
0.998839,
0.999157,
0.999389,
0.999558,
0.999679,
0.999936,
0.999987,
0.999997,
0.999999,
0.999999,
0.999999,
0.999999,
0.999999
]
}
SM1 FIT
false
SM2 FIT
false
SM3 FIT
false
SM4 FIT
false
SM5 FIT
false
Figures